| Research Article | ||
J. Microbiol. Infect. Dis., (2024), Vol. 14(2): 67–79 Research Article Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri LankaHewa Babarandage Chathurika Harshani1*, Dilshani Kavindya Balasuriya2, Wickramasinghe Widana Pathirana Pragathi Nirasha Diwyanjalee2, Aranwela Gamage Gaya Kaushalya1 and Wijendra Acharige Somalatha Wijendra11Medical Research Institute, Colombo, Sri Lanka 2Sri Lanka Institute of Information Technology, Malabe, Sri Lanka *Corresponding Author: Hewa Babarandage Chathurika Harshani. Medical Research Institute, Colombo, Sri Lanka. Email: hbc.harshani [at] gmail.com Submitted: 14/03/2024 Accepted: 20/06/2024 Published: 30/06/2024 © 2024 Journal of Microbiology and Infectious Diseases
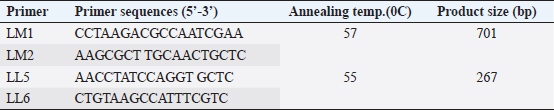
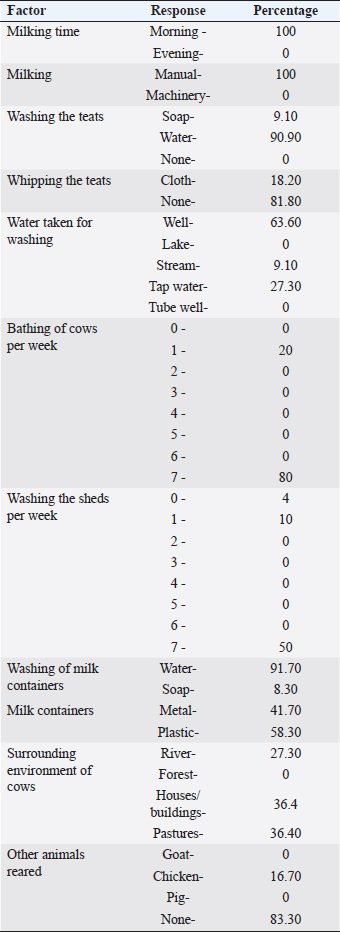
AbstractBackground: Listeria monocytogenes, a foodborne pathogen, poses a significant risk to public health due to its potential to cause severe infections termed “listeriosis”. Raw milk and dairy products are common vehicles for L. monocytogenes transmission, amplifying the risk to consumers. Additionally, the emergence of antibiotic resistance in Listeria strains heightens concerns in human and veterinary medicine. Aim: This study aimed to assess the prevalence of L. monocytogenes in raw milk samples from Sri Lanka’s Colombo district and to characterize the antibiotic resistance patterns of positive isolates. Methods: One hundred raw milk samples were collected from various milk collection centers. Samples were sourced from locations including Awissawella, Kahanthota, Meepe, Homagama, and Udahamulla. Antibiotic resistance was evaluated using standard methods like disk diffusion and well diffusion, testing ampicillin, penicillin, streptomycin, and tetracycline. Results: Of the 100 samples, L. monocytogenes was detected by conventional PCR in 83, with Awissawella having the highest occurrence (100%), followed by Kahanthota (81.81%), Meepe (81.48%), Homagama (77.27%), and Udahamulla (44.44%). Among the positive samples, 72 showed varying degrees of antibiotic resistance. The highest level of resistance was observed for penicillin (83.1%), followed by ampicillin (65%), streptomycin (32.53%), and tetracycline (10.84%). Sixty isolates (60%) displayed resistance to more than one antibiotic studied. Findings also revealed the widespread manual milking, predominant water usage over soap for hygiene, and common daily bathing of cows, highlighting the need for improved hygiene practices to minimize contamination risks in dairy farming. Conclusion: Raw milk contaminated with L. monocytogenes pose a serious public health risk. The study highlights the need for stringent hygiene measures in milk production and processing to safeguard consumers. Additionally, the high prevalence of antibiotic-resistant strains underscores the importance of judicious antibiotic use in both human and veterinary medicine to mitigate the further emergence and spread of resistant Listeria strains. Keywords: Antibiotic resistance, Listeria monocytogenes, Raw milk, PCR, Antibiotic Susceptibility Testing. IntroductionListeria monocytogenes is a microaerophilic, Gram-positive pathogen, which is a member of the Listeriaceae family comprising 20 species (Nwaiwu, 2020). Among them, L. monocytogenes is the sole pathogenic species, posing significant risks to both human and animal health. It is very adaptable to a wide range of environmental settings, including acidity (pH) between 4.3 and 9.6, water activity at approximately 0.83, and salt concentrations of up to 10% (Shamloo et al., 2019). Listeria are also able to proliferate in low-temperature environments such as fridges where most foods are believed to be safe from most pathogens (Osek et al., 2022). As a result, it is ubiquitously present in diverse ecosystems, including soil, water, and a variety of food products. Listeria can establish chronic infections and persist in varied conditions within the digestive system (Gahan et al., 2014). The infection induced by L. monocytogenes is referred to as listeriosis, and is primarily induced through the ingesting of infected foods (Liu et al., 2005). Listeriosis outbreaks have a notably high fatality rate, ranging from 20% to 30%. This fatality rate is significantly higher when compared to other common foodborne diseases like Salmonella enteritidis (0.38% mortality), Vibrio spp. (0.005%–0.01% mortality), and Campylobacter spp (0.02%–0.1% mortality) (EFSA and ECDC, 2021). Listeriosis is a disease with severe health implications. It comprises two forms: non-invasive and invasive. Non-invasive listeriosis is a milder form of this illness that typically impacts healthy individuals. Symptoms include diarrhea, muscle ache, fever, and headache, with a relatively brief incubation time lasting only a few days. In contrast, invasive listeriosis represents a more fatal variant of the condition, primarily impacting vulnerable groups such as pregnant mothers, individuals with cancer, those with HIV, organ transplant recipients, the elderly, and newborns (Hilliard et al., 2018). Common symptoms consist of muscle ache, fever, septicemia, and meningitis, with a longer incubation period of 1–2 weeks. Moreover, L. monocytogenes outbreaks are often linked to contaminated dairy products resulting in fatalities and substantial economic losses. Raw milk has been linked to recorded incidences of contamination caused by mastitis in lactating cows as a potential source of L. monocytogenes infection. Despite the ability of pasteurization to lower the risk of infection, incidents of milk contamination have been reported, emphasizing that the threat cannot be completely eliminated even after pasteurization (Lyytikäinen et al., 2006). This situation poses a significant global challenge, particularly in developing countries like Sri Lanka. Beyond the threat posed by L. monocytogenes, antibiotic resistance has become a pressing global concern. The extensive usage of antibiotics has played a major role in developing resistance to antibiotics in humans as well as animals (Imperial and Ibana, 2016; Agyare et al., 2018). When a bacterial pathogen becomes resistant to antibacterial therapy, it can delay the administration of appropriate treatment, and the antibiotics needed to combat resistant pathogens may lose their effectiveness or become toxic. It is estimated that overall deaths caused due to antibiotic-resistant bacterial 17 infections could surge between 700,000 and 10 million every year, with associated expenses exceeding $100 trillion by 2050 (Olaimat et al., 2018). Listeria species display inherent resistance to specific antibiotics, such as fosfomycin, first-generation quinolones, cephalosporin, and sulfamethoxazole (Troxler et al., 2000; Conter et al., 2009). The emergence of antimicrobial resistance in L. monocytogenes pose a growing concern, particularly within the food sector (Buchanan et al.,2017; Gelbíčová et al., 2018; Aureli et al., 2000; Keet & Rip, 2021; WHO, 2024). Even more concerning is the appearance of L. monocytogenes which are resistant to multiple drugs, which make common antibiotics ineffective (Manyi-Loh et al., 2023). In this study, we aim to detect antibiotic resistance of L. monocytogenes against 4 antibiotics; penicillin, ampicillin, streptomycin, and tetracycline in raw milk samples of Colombo district, Sri Lanka. The above antibiotics are widely used in cattle farms to treat livestock in the selected areas. Not only that, some of them are (Penicillin, ampicillin, and amoxicillin) frequently used for treating Listeria infections (Ishihara and Akazawa, 2023). Materials and MethodsSample collectionIn this study, samples were collected from randomly selected milk collection centers across the geographical location using a convenient sampling method. These samples were collected from June 2023 to September 2023. Five milk collection centers within the Colombo district were selected at random and a total of one hundred raw milk samples were collected. These one hundred samples were collected from five milk collection centers in the areas; Awissawella (31), Homagama (22), Meepe (27), Udahamulla (09), Kahanthota (11). The samples were then brought to the laboratory in cold conditions and stored at 40C. Description of milk collection centersThe milk collection centers, strategically located near farmers’ residences, were operated from 8.00 to 9.00 am to facilitate timely milk transfer. Milk was delivered by farmers in aluminum or plastic cans, which were weighed upon arrival by designated officers. The recorded weights were used to aid in inventory management. Subsequently, the milk was carefully poured into collection tanks, often equipped with strainers to maintain purity by filtering out impurities or debris. Quality checks were conducted, focusing on antibiotic residues such as Penicillin and tetracyclines, farm-level hygiene, and overall milk cleanliness. Farm-level data were deemed crucial for assessing contamination risks, notably from Listeria strains, and understanding transportation conditions. The number of participating farmers varied, with an average of 30–40 farmers per center during the morning collection period. Stringent quality control measures were implemented to ensure milk integrity and consumer safety throughout the process. IsolationThe samples were enriched in Listeria Selective Enrichment Broth and incubated at 37°C for 48 hours. The enriched samples were then subjected to spread plating on Listeria Selective Agar (Oxoid-UK) plates and incubated at 37°C for 48 hours. After incubation, the morphological characteristics of the bacterial colonies were examined. Among the colonies, those exhibiting features that closely resemble the characteristics typically observed in Listeria spp. colonies, such as a blackish–brown color surrounding the colonies, 0.5 mm diameter, ash-colored, circular, convex, and entirely edged colonies, were singled out and further isolated to obtain discrete colonies. DNA extractionDNA extraction of the bacterial colonies was done using the Wizard Genomic DNA Purification kit (Promega). The DNA was quantified to determine its concentration in the NanoDrop™ 1000 spectrophotometer. Detection of L. monocytogenesThe isolates of L. monocytogenes were confirmed by a two-step nested PCR amplification using primers targeting the ‘Listeriolysin O’ gene by using a modified method previously stated by Herman et al. 1995. The initial PCR utilized the primers, LM1 (F) and LM2 (R). The second PCR utilized nested primers LL5 and LL6 (Table 1). The first PCR was performed in a final volume of 50 μl, including 10 μl of PCR buffer (5×), 5 μl of MgCl2 (25 mM), 1 μl of a dNTPs mix (10 mM), 0.25 μl of Taq polymerase (5U/μl), and 50 pmol of both forward and reverse primers. In the initial PCR, 10 μl of the sample DNA was introduced. The PCR reaction was performed in a Bio-Rad PCR System (MYCyclerTM thermal cycler). The cycling conditions included an initial denaturation at 95°C for 2 minutes, followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 57°C for 30 seconds, and an elongation step at 72°C for 60 seconds. Final elongation was carried out at 72°C for 5 minutes. For the nested PCR, the template consisted of 10 μl of the first PCR product diluted at a ratio of 1:10 while keeping the other reagents consistent as the initial PCR. The second PCR was conducted with the cycling conditions including an initial denaturation at 95°C for 2 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and elongation at 72°C for 60 seconds. Final elongation was carried out at 72°C for 5 minutes. Finally, the PCR product was analyzed by agarose gel electrophoresis with a 2% gel and Trisborate-EDTA buffer system. The PCR product was visualized under the UV transilluminator after staining with ethidium bromide for 5 minutes. The expected size of the final PCR product was 267 base pairs. Antibiotic susceptibility testingAntibiotic susceptibility testing was conducted on bacterial isolates that was tested positive for Listeria monocytogenes using four different antibiotic disks: penicillin-G (10IU), ampicillin (10 µg), streptomycin (10 µg), and tetracycline (30 µg). To perform the tests, swabs from bacterial suspensions, equivalent to the standard of 0.5 McFarland. (approximately 1 × 108 CFU/ml), were spread onto Mueller-Hinton Agar plates (OXOID, UK). Subsequently, the antibiotic disks were dispensed on the surface of the agar plates containing the bacterial cultures. In instances when the standard antibiotic disks were not accessible, the experiment proceeded by employing a modified version of the well diffusion assay method outlined by Holder and Boyce (1994). After an incubation period of 24 hours at 37°C, measurements were taken for the diameter of the inhibition zone surrounding each antibiotic disk. The experiment was triplicated to minimize potential errors. The results were then categorized as resistant, intermediate, or susceptible based on the parameters outlined by the Clinical and Laboratory Standards Institute (CLSI, 2020), as outlined in Table 2. The study relied on the breakpoints established for Staphylococcus spp. because the breakpoints specific for Listeria spp. was not available in the CLSI guidelines (Chen et al., 2018; Faridi et al., 2021). If a bacterium displayed resistance to more than one antibiotic, it was considered as exhibiting resistance to multiple antibiotics used in this study. The study was also encompassed an analysis of factors influencing farmers’ milking practices, drawing insights from data collected from farmers bringing milk to milk collection centers. Table 1. Primers used in detecting L. monocytogenes (Harshani et al., 2022).
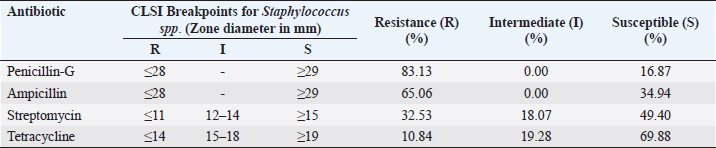
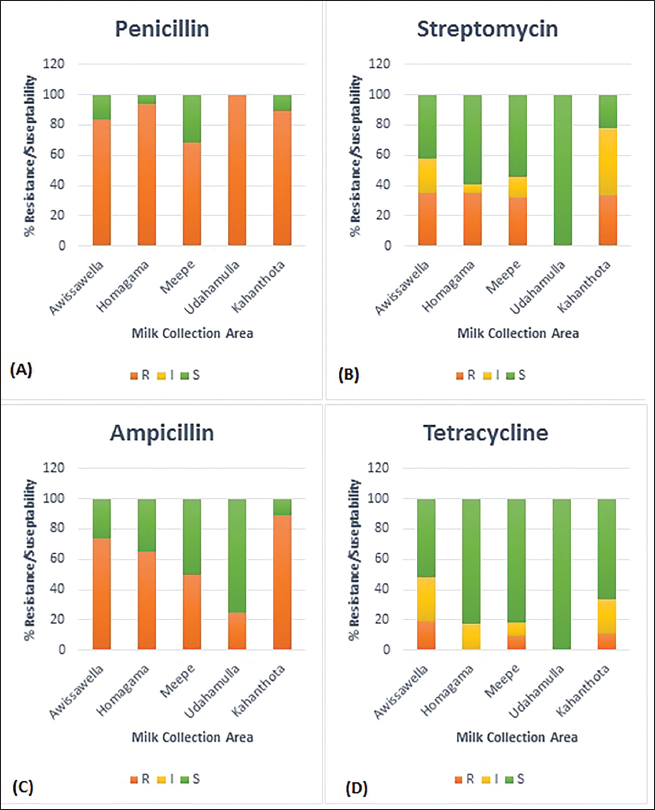
Table 2. Percentage of resistance (R), intermediate resistance (I), and susceptible(S) for four distinct antibiotics related to L. monocytogenes samples from the Colombo District.
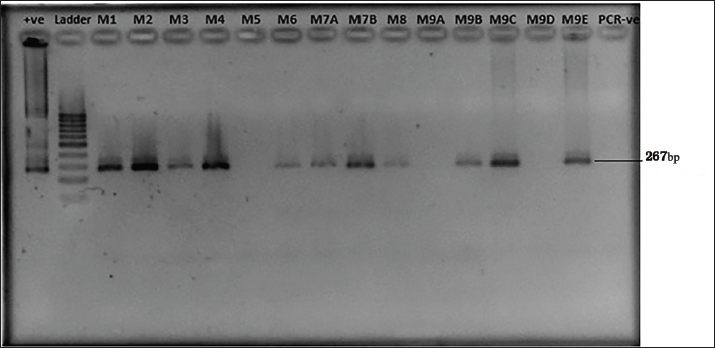
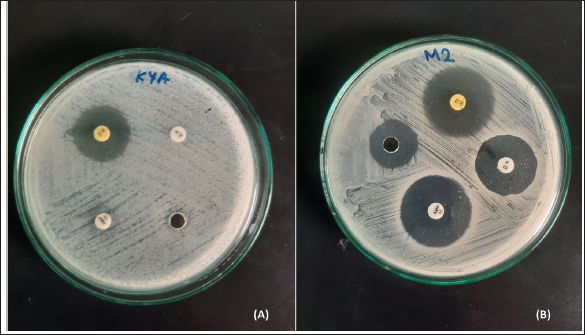
Ethical approvalEthics approval was granted by the Ethics Review Committee, Medical Research Institute Colombo, Sri Lanka Project No: ERC/MRI/03/2023. ResultsPrevalence and detection of L. monocytogenesMany isolates appeared as creamish yellow, circular, convex colonies with varying colony sizes from 0.5 mm to 2 mm in diameter resembling the positive control ( L. monocytogenes ATCC 51776). The characteristic black color zone around colonies indicated the potential existence of L. monocytogenes among these samples. The presence of L. monocytogenes was confirmed by PCR. Out of the 100 samples tested, 83 were identified as positive by the presence of a PCR amplicon of 267bp (Fig. 1; Supplementary Figs. 1–5). Antibiotic susceptibility testAntibiotic resistance profiles were determined for 83 individual L. monocytogenes isolates, focusing on specific antibiotics. Diverse patterns of antibiotic resistance were observed among these isolates, with variations in resistance to penicillin, ampicillin, and tetracycline as determined by standard disk diffusion assays, and streptomycin as assessed using well diffusion assays as shown in Figure 2. Among the positive L. monocytogenes isolates, 83.13% (69/83) demonstrated resistance to penicillin, making it the antibiotic to which the bacteria exhibited the highest resistance. Conversely, the lowest resistance rate was observed for tetracycline, with only 10.84% (9/83) of isolates showing resistance. Intermediate resistance was observed in 18.07% (15/83) of cases for streptomycin and in 19.27% (16/83) for tetracycline. Among the examined strains of L. monocytogenes, 69.88% (58/83) showed susceptibility to tetracycline, making it the antibiotic with the highest percentage of susceptible strains, while only 16.87% (14/83) exhibited susceptibility to penicillin, indicating it had the lowest percentage of susceptible strains. Overall, the majority of isolates (86.75%) demonstrated resistance to each antibiotic tested, while only a few displayed intermediate resistance or susceptibility to the concentrations used in the testing (Table 2). As indicated by the phenotypic characterization results in this study, among the 72 resistant isolates, 12 (41.2%) were resistant to only one antibiotic (Penicillin-10, Ampicillin-1, Streptomycin-1, and Tetracycline-0). Additionally, 35 isolates demonstrated resistance to a minimum of two antibiotics (Penicillin and Ampicillin – 28; Penicillin and Tetracycline – 01; Penicillin and Streptomycin – 05; Streptomycin and Tetracycline – 01), while another 23 isolates displayed resistance to at least three antibiotics (Penicillin, Ampicillin, Streptomycin – 18; Penicillin, Ampicillin, Tetracycline – 05). Interestingly, 04 isolates demonstrated resistance against all four antibiotics. These results highlight the existence of isolates with resistance to multiple antibiotics in these areas. The percentage of antibacterial susceptibility in positive isolates varied among the four geographic regions for the four distinct antibiotics (Fig. 3). Resistance to penicillin was evident in all five locations, with Awissawella 26/31(83.8%), Homagama 16/17(94.1%), Meepe 15/22(68.2%), Udahamulla 4/4(100%), and Kahanthota 8/9(88.9%) showing notable resistance. Similarly, ampicillin resistance was prevalent across all areas, with resistance percentages as follows: Awissawella 23/31(74.2%), Homagama 11/17(64.7%), Meepe 11/22(50%), Udahamulla 1/4(25%), and Kahanthota 8/9(88.9%). Tetracycline resistance was prevalent only in few areas including; Awissawella 6/31(19.4%), Meepe 2/22(9.09%), and Kahanthota 1/9(11.11%). As for streptomycin resistance, it was observed in Awissawella 11/31(35.5%), Homagama 6/17(35.3%), Meepe 7/22(31.8%), and Kahanthota 3/9(33.3%). Udahamulla had the highest resistance to penicillin-G, while Meepe exhibited the lowest resistance. Kahanthota displayed the highest resistance to ampicillin, while Udahamulla showed the lowest resistance. Awissawella had the highest resistance percentages for both streptomycin and tetracycline. While Udahamulla exhibited the least resistance to streptomycin and tetracycline. In comparison to penicillin-G, streptomycin, and ampicillin, the resistance against tetracycline was notably lower. Similarly, isolates from Udahamulla and Meepe demonstrated lower resistance to all four antibiotics compared to the other three areas, while isolates from Awissawella and Kahanthota exhibited higher resistance to all four antibiotics in comparison to the other areas.
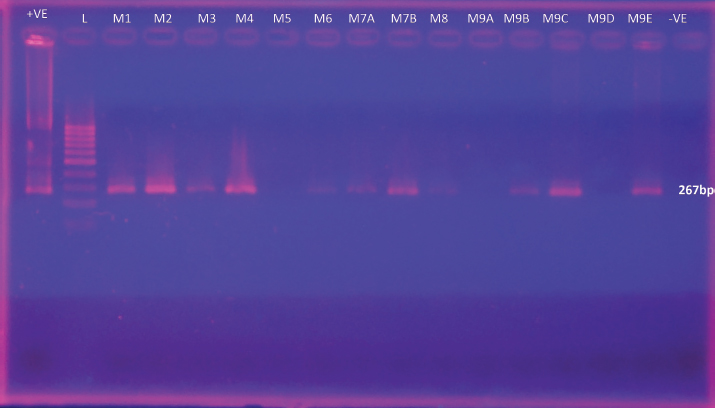
Fig. 1. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lanes M1 to M10 correspond to samples from Meepe 1 to Meepe 10 respectively and the lane PCR -ve represents the negative control for PCR with Milli-Q water.
Fig. 2. Listeria monocytogenes isolate from (A)—K4A sample, (B)—M2 sample on Mueller Hinton agar with antibiotic disks (A) Penicilin—G, (B) Ampicilin, (C) Tetracycline, (D) Streptomycin. Factors influencing farmers’ milking practicesInformation regarding dairy farming practices from the farmers who carry milk for the five milk collection centers were also analyzed in this study (Table 3). Manual milking is universally practiced (100%), while mechanical milking remains unused (0%). Hygiene standards during milking show a predominance of water usage (90.90%) over soap (9.10%). Daily bathing of cows is common (80%), and sheds are typically washed weekly (50%). Wells (63.60%) and tap water (27.30%) are the primary sources for washing, indicating accessibility and quality as important factors. Cows predominantly graze in pastures (36.40%), with a notable presence near rivers (27.30%), highlighting potential environmental contamination risks. These results underscore the necessity for targeted interventions to enhance hygiene practices and minimize contamination risks in dairy farming. DiscussionThe widespread use of antibiotics in recent years has led to occasional misuse in both humans and animals. Therefore, this plays a vital role in the development and distribution of resistance to antibiotics among foodborne bacteria such as L. monocytogenes. Notably, antibiotic consumption has been substantial in Sri Lanka, as evidenced by a survey conducted by Sri Ranganathan et al. in (2021), which reported a high antibiotic consumption rate and the disproportionate use of broad-spectrum antibiotics. Antibiotics are also widely employed in animal husbandry to prevent infections and promote growth. In Sri Lanka, poultry and livestock farms serve as reservoirs of antibiotic-resistant bacteria, with commonly used antibiotics including tetracycline, ampicillin, oxytetracycline, and penicillin (Liyanage and Pathmalal, 2017). Although L. monocytogenes can be transmitted through various food sources, the contamination of milk and milk products with is particularly critical globally, especially in developing countries like Sri Lanka. Despite the severity of this challenge, only a handful of reports have investigated the presence of L. monocytogenes in dairy products in Sri Lanka. Previous studies have highlighted higher percentages of L. monocytogenes-contaminated milk samples (Wijesundera et al., 1992; Jayamanne and Samarajeewa, 2001; Wijendra et al., 2014; Harshani et al., 2022).
Fig. 3. The percentage of antibacterial susceptibility of positive isolates from different areas to four different antibiotics; (A) Penicillin—G, (B) Streptomycin, (C) Ampicillin, (D) Tetracycline. Our study revealed diverse and notable patterns of antibiotic susceptibility profiles of L. monocytogenes isolates in raw milk of Colombo District. High resistance rates were observed against penicillin (83.13%) followed by ampicillin (65.06%), streptomycin (32.53%), and tetracycline (10.84%). These results resonate with previous studies, underlining the challenge posed by elevated resistance to specific antibiotics. The identification of intermediate resistance for streptomycin (18.07%) and tetracycline (19.28%) suggests a potential towards the gradual development of resistance against these antibiotics in L. monocytogenes. Previous studies have shown that L. monocytogenes derived from both food and animals can be resistant to a variety of medicines including, streptomycin, chloramphenicol, gentamicin, erythromycin, rifampin, and kanamycin (Charpentier et al., 1995; Walsh et al., 2001). Research conducted in the Pollonaruwa District revealed higher resistance levels for ampicillin (60%) with lower resistance for tetracycline (8%), with 40% of isolates exhibiting phenotypic multiple antibiotic resistance (Harshani et al., 2022). Based on the disk diffusion assay, our own research findings revealed higher resistance to penicillin and the lowest resistance to tetracycline. Table 3. Summary of factors leading to milk contamination in cattle farms.
In the current study, conventional PCR was carried out for the identification of L. monocytogenes. The authors chose two-step nested PCR amplification to confirm the presence of the Listeriolysin O (LLO) gene in L. monocytogenes due to its high specificity and sensitivity. Nested PCR involves two sets of primers: the first set amplifies a larger region of DNA containing the target gene, and the second set amplifies a smaller region within the product of the first PCR. This increases the specificity of the amplification, reducing the likelihood of non-specific amplification. Targeting the LLO gene specifically is crucial because it is a virulence factor associated with L. monocytogenes pathogenicity. Confirming its presence helps in identifying the pathogenic potential of the strain being studied. The high rate of contamination of L. monocytogenes in raw milk might be due to poor sanitation practices among milking cows and due to corn silage and other cow feed being contaminated with L. monocytogenes, which could infect cows through ingestion (Husu et al., 1990). Another aspect of udder cleanliness is drying the udders with a cloth before milking. Failure to do so may result in bacterial cells being washed into the milk by water running down the udder skin into the milk container (Moroni et al., 2018). Therefore, drying the udders with a cloth before milking is important for avoiding or minimizing contamination. The source of water used for washing cows and sheds is also crucial. Ground and surface water resources are commonly used, but they may be contaminated with pathogenic bacteria, increasing the risk of contamination. Additionally, the use of tanks for washing cows may contribute to L. monocytogenes contamination, as this bacterium often inhabits moist soil and water (Singh et al., 2020). Finally, the cleanliness of milking equipment, particularly the way milk containers are washed, is vital. While most farmers use soap to clean containers, a small percentage relies solely on water for cleaning. The combined influence of these factors leads to the contamination of L. monocytogenes in raw milk, emphasizing the need to improve sanitation practices among milking cows and their environments (Husu et al., 1990). The analysis several critical factors that could influence the risk of L. monocytogenes contamination in raw milk were also analyzed in this study. Manual milking, which is practiced by 100% of farmers surveyed, may pose a higher risk compared to mechanical milking, as it involves more direct contact between the milker and the animal, potentially introducing contaminants. Furthermore, the relatively low usage of soap (9.10%) during teat washing suggests potential shortcomings in cleaning practices, which could contribute to the persistence of L. monocytogenes. Additionally, the frequency of bathing cows and washing sheds per week is a significant factor, as inadequate cleaning may lead to the buildup of L. monocytogenes in the environment. The choice of water source for washing is also crucial, particularly if sourced from rivers (27.30%) or wells (63.60%), as these may be susceptible to environmental contamination. Overall, these findings underscore the importance of implementing stringent hygiene protocols, including thorough teat washing with effective disinfectants, regular cleaning of cow sheds and equipment, and ensuring the quality and safety of water sources to minimize the risk of L. monocytogenes contamination in raw milk. The comprehensive examination of antibiotic use, antibiotic resistance patterns, and factors influencing L. monocytogenes contamination in raw milk underscores the complexity of ensuring food safety and public health in dairy production. The widespread use and misuse of antibiotics, coupled with inadequate sanitation practices and environmental contamination, pose significant challenges in mitigating the risk of L. monocytogenes contamination in raw milk. Addressing these multifaceted issues requires collaborative efforts among stakeholders, including policymakers, farmers, healthcare professionals, and researchers. Implementing stringent hygiene protocols, promoting responsible antibiotic use in both human and animal healthcare, and enhancing surveillance and monitoring systems are imperative steps towards minimizing the risk of L. monocytogenes contamination in raw milk. By prioritizing these measures, we can safeguard the integrity of our food supply chain and protect consumers from potential health hazards associated with L. monocytogenes contamination in dairy products. ConclusionIn conclusion, the comprehensive analysis of dairy farming practices, antibiotic use, antibiotic resistance patterns, and factors influencing L. monocytogenes contamination in raw milk underscores the multifaceted nature of ensuring food safety and public health in dairy production. The widespread use and misuse of antibiotics, coupled with inadequate sanitation practices and environmental contamination, pose significant challenges in mitigating the risk of L. monocytogenes contamination in raw milk. Addressing these challenges requires concerted efforts from various stakeholders, including policymakers, farmers, healthcare professionals, and researchers. Implementing stringent hygiene protocols, promoting responsible antibiotic use, enhancing surveillance and monitoring systems, and improving sanitation practices are crucial steps towards minimizing the risk of L. monocytogenes contamination in raw milk. By prioritizing these measures, we can safeguard the integrity of our food supply chain and protect consumers from potential health hazards associated with L. monocytogenes contamination in dairy products. AcknowledgmentThe authors would like to express their sincere gratitude to the Department of Molecular Biology, Medical Research Institute, Sri Lanka. The authors are also thankful to Medical Research Institute and Ministry of Health, Sri Lanka for the financial support provided. FundingThis study is funded by the Research Committee, Medical Research Institute Colombo, Sri Lanka, Grant No: RC/MRI/02/2023. Authors’ contributionHBCH and WASW led conceptualization, design, intellectual content, supervision and review, and editing of final manuscript. DKB & WWPPND contributed to the literature search, experimental studies and data analysis. AGGK contributed to the experimental studies. HBCH, WASW, DKB, WWPPND & AGGK contributed to the manuscript preparation. Conflict of interestAll authors declare there is no conflict of interest. Data availabilityThe data that support the findings of this study are available on request from the corresponding author. ReferencesAgyare, C., Boamah, V.E., Zumbi, C.N. and Osei FB. 2018. Antibiotic use in poultry production and its effects on bacterial resistance. Antimicrob Resist-A Global Threat. 33–50. doi: http://doi.org/10.5772/intechopen.79371 Aureli, P., Fiorucci, G.C., Caroli, D., Marchiaro, G., Novara, O., Leone, L. and Salmaso S. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med. 342, 1236–1241. Buchanan, R.L., Gorris, L.G.M., Hayman, M.M., Jackson T.C., and Whiting R.C. 2017. A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 75, 1–13. Charpentier, E., Gerbaud, G., Jacquet, C., Rocourt, J and Courvalin P. 1995. Incidence of antibiotic resistance in Listeria species. J Infect Dis.172(1), 277–281. doi: http://doi.org/10.1093/infdis/172.1.277 Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Xue, L., Lei, T., Zeng, H., Wu, S., Ye, Q., Bai, J and Wang J. 2018. Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes isolated from fresh aquatic products in China. Front Microbiol. 9, 2215. doi: http://doi.org/10.3389/fmicb.2018.02215 CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100.Wayne, PA: Clinical and Laboratory Standards Institute. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf Conter, M., Paludi, D., Zanardi, E., Ghidini, S., Vergara, A. and Ianieri A. 2009. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int J Food Microbiol. 128(3), 497–500. doi: http://doi.org/10.1016/j.ijfoodmicro.2008.10.018EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2021. The European Union One Health 2020 Zoonoses Report. EFSA. J. 19(12), 6971, 324. Available via https://www.efsa.europa.eu/en/efsajournal/pub/7666 Faridi, Z., Soltan Dallal, MM., Koohy-Kamaly, P., Haji Seyed Javadi, N., Hadian, Z. and Mazaheri Nezhad Fard R. 2021. Prevalence and antimicrobial resistance of Listeria monocytogenes in raw milk in Tehran, Iran. Appl Food Biotechnol. 8(3), 181–188. Available via https://journals.sbmu.ac.ir/afb/article/view/34379 Gahan Cormac, GM. and Hill C. 2014. “Listeria monocytogenes: survival and adaptation in the gastrointestinal tract”. Front. Cell. Infect. Microbiol. 4(9), 5. doi: http://doi.org/10.3389/fcimb.2014.00009 Gelbíčová, T., Zobaníková, M., Tomáštíková, Z., Van Walle, I., Ruppitsch, W. and Karpíšková, R. 2018. An outbreak of listeriosis linked to turkey meat products in the Czech Republic, 2012–2016. Epidemiol Infect. 146, 1407–1412. Harshani, H.B.C., Ramesh, R., Halmillawewa, A.P. and Wijendra, W.A.S. 2022. Phenotypic and genotypic characterization of antibiotic resistance of Listeria monocytogenes isolated from raw milk samples collected from Polonnaruwa, Sri Lanka. EJFA. 34(1), 16. doi: http://doi.org/10.9755/ejfa. 2022.v34.i1.2809 Herman, L.M, De Block, J.H. and Moermans R.J. 1995. Direct detection of Listeria monocytogenes in 25 milliliters of raw milk by a two-step PCR with nested primers. Appl. Environ. Microbiol. 61(2), 817–819. doi: http://doi.org/10.1128/aem.61.2.817-819.1995 Hilliard, A., Leong D. O’Callaghan, A., Culligan, EP., Morgan, CA., DeLappe, N., Hill, C., Jordan, K Cormican, M. and Gahan, CGM. 2018. Genomic characterization of Listeria monocytogenes isolates associated with clinical Listeriosis and food production environment in Ireland. Genes. 9, 171. doi: http://doi.org/10.3390/genes9030171 Holder, I.A. and Boyce, S.T. 1994. Agar well diffusion assay testing of bacterial susceptibility to various antimicrobials in concentrations non-toxic for human cells in culture. Burns. 20(5), 426–429. doi: http://doi.org/10.1016/0305-4179(94)90035-3 Husu, J.R, Seppänen, J.T, Sivelä, S.K. and Rauramaa, AL. 1990. Contamination of raw milk by Listeria monocytogenes on dairy farms. Zentralblatt fur Veterinarmedizin. Reihe B. J. Vet. Med. Series B. 37(4), 268–275. doi: http://doi.org/10.1111/j.1439-0450. 1990.tb01058.x Imperial, IC. and Ibana, J A. 2016. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front. Microbiol. 7, 1983. doi: http://doi.org/10.3389/fmicb.2016.01983 Ishihara, Y. and Akazawa, K. 2023. Treatment of Listeria monocytogenes bacteremia with oral levofloxacin in an immunocompromised patient. IDCases. 31, e01680. doi: http://doi.org/10.1016/j.idcr.2023.e01680 Jayamanne, VS. and Samarajeewa U. 2001. Incidence and detection of Listeria monocytogenes in milk and milk products of Sri Lanka. Trop. Agric. Res. 13, 42–50. doi: http://doi.org/10.4038/tare.v13i3.3142 Keet, R. and Rip, D. 2021. Listeria monocytogenes isolates from Western Cape, South Africa exhibit resistance to multiple antibiotics and contradicts certain global resistance patterns. AIMS Microbiol. 7(1), 40–58. doi: http://doi.org/10.3934/microbiol.2021004 Liu, D., Lawrence, M.L., Ainsworth, A.J. and Austin, F.W. 2005. Isolation and PCR amplification of a species-specific oxidoreductase-coding gene region in Can. J. Microbiol. 51(1), 95–98. doi: http://doi.org/10.1139/w04-108 Liyanage, G.Y. and Pathmalal, M. 2017. Risk of prophylactic antibiotics in livestock and poultry farms: a growing problem for human and animal health. Pharm. J. Sri Lanka. 7(1), 13–22. doi: http://doi.org/10.4038/pjsl.v7i0.16 Lyytikäinen, O., Nakari, U.M., Lukinmaa, S., Kela, E., Minh, N.N.T. and Siitonen, A. 2006. Surveillance of listeriosis in Finland during 1995-2004. Eurosurveillance. 11(6), 5–6. Available via https://pubmed.ncbi.nlm.nih.gov/16801696 Manyi-Loh, C.E., Okoh, A.I., and Lues, R. 2023. Occurrence and multidrug resistance in strains of Listeria monocytogenes recovered from the anaerobic co-digestion sludge contained in a single stage steel biodigester: implications for antimicrobial stewardship. Microorganisms. 11(3), 725. doi: http://doi.org/10.3390/microorganisms11030725 Nwaiwu O. 2020. What are the recognized species of the genus Listeria? Access Microbiol. 2(9), acmi000153. doi: http://doi.org/10.1099/acmi.0.000153 Olaimat, A.N., Al-Holy, M A., Shahbaz, H.M., Al-Nabulsi, A.A., Abu Ghoush, M.H., Osaili, T.M. and Holley, R.A. 2018. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. CRFSFS. 17(5), 1277–1292. doi: http://doi.org/10.1111/1541-4337.12387 Osek, J., Lachtara, B. and Wieczorek, K. 2022. Listeria monocytogenes—How this pathogen survives in food-production environments? Front Microbiol. 13, 866462. doi: http://doi.org/10.3389/fmicb.2022.866462 Shamloo, E., Hosseini, H., Moghadam, Z.A., Larsen, M.H., Haslberger, A. and Alebouyeh, M. 2019. Importance of Listeria monocytogenes in food safety: a review of its prevalence, detection, and antibiotic resistance. Iran. J. Vet. Res. 20(4), 241. Singh, A., and Ramachandran, A. 2020. “Assessment of Hygienic Milking Practices and Prevalence of Bovine Mastitis in Small Dairy Farms of Peri-Urban Area of Jaipur.” Indian J Community Med. 45(Suppl 1), S21–S25. doi: http://doi.org/10.4103/ijcm.IJCM_363_19 Sri Ranganathan, S., Wanigatunge, C., Senadheera, G.P.S.G. and Beneragama, B.V.S.H. 2021. A national survey of antibacterial consumption in Sri Lanka. PLoS One. 16(9):e0257424. doi: http://doi.org/10.1371/journal.pone.0257424 Stefan, G. and Baraitareanu, S. 2023Approaches of milking biosecurity and milking parlour hygiene in dairy farms. Recent developments on mastitis—treatment and control. London, UK: IntechOpen. Available via http://dx.doi.org/10.5772/intechopen.113084 Troxler, R., von Graevenitz, A., Funke, G., Wiedemann, B. and Stock, I. 2000. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6(10), 525–535. doi: http://doi.org/10.1046/j.1469-0691.2000. 00168.x Walsh, D., Duffy, G., Sheridan, J.J., Blair, I.S. and McDowell D.A. 2001. Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J. Appl. Microbiol. 90(4), 517–522. doi: http://doi.org/10.1046/j.1365-2672.2001.01273.x Wijendra, W.A.S., Kulathunga, K.A.K. and Ramesh, R. 2014. First report of Listeria monocytogenes serotypes detected from milk and milk products in Sri Lanka. Adv. Anim. Vet. Sci. 2(5S), 11–16. doi: http://doi.org/10.14737/journal.aavs/2014/2.5s.11.16 Wijesundera, C.D., Pinto, M.R. and Navaratnam, C. 1992. Two cases of listeriosis. Cey. Med. 37, 21–23. Available via https://pubmed.ncbi.nlm.nih.gov/1581992 World Health Organization. 2024. Antimicrobial resistance. Available via https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
Supplementary Fig. 1. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lane L- Ladder, Lanes A1 to A14 correspond to samples from Awissawella 1 to Awissawella 14 respectively and the lane PCR −ve represents the negative control for PCR with Milli-Q water.
Supplementary Fig. 2. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lane L- Ladder, Lanes A15 to A28 correspond to samples from Awissawella 15 to Awissawella 28 respectively and the lane PCR −ve represents the negative.
Supplementary Fig. 3. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lane L- Ladder, Lanes H1 to H12 correspond to samples from Homagama 1 to Homagama 12 respectively and the lane PCR −ve represents the negative.
Supplementary Fig. 4. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lane L- Ladder, Lanes M1 to M9E correspond to samples from Meepe 1 to Meepe 9E respectively and the lane PCR −ve represents the negative.
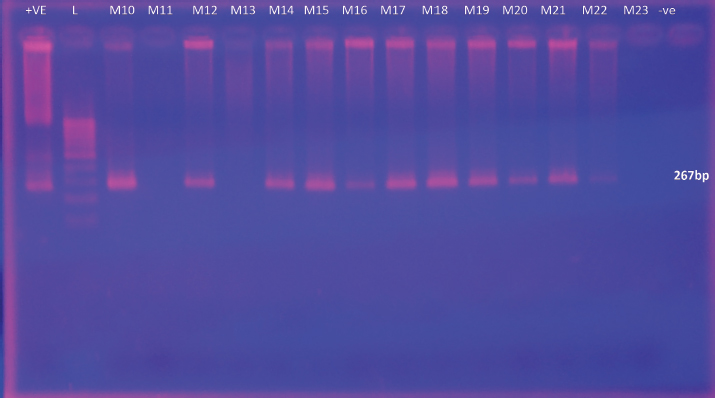
Supplementary Fig. 5. Gel Image for detection of L. monocytogenes by nested PCR. Lane +ve represents L. monocytogenes ATCC 51776 (the positive control for the target gene), Lane L- Ladder, Lanes M10 to M23 correspond to samples from Meepe 10 to Meepe 23 respectively and the lane PCR −ve represents the negative | ||
| How to Cite this Article |
| Pubmed Style Harshani HBC, Balasuriya DK, , Kaushalya AGG, Wijendra WAS. Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. J Microbiol Infect Dis. 2024; 14(2): 67-79. doi:10.5455/JMID.2024.v14.i2.5 Web Style Harshani HBC, Balasuriya DK, , Kaushalya AGG, Wijendra WAS. Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. https://www.jmidonline.org/?mno=194248 [Access: January 03, 2026]. doi:10.5455/JMID.2024.v14.i2.5 AMA (American Medical Association) Style Harshani HBC, Balasuriya DK, , Kaushalya AGG, Wijendra WAS. Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. J Microbiol Infect Dis. 2024; 14(2): 67-79. doi:10.5455/JMID.2024.v14.i2.5 Vancouver/ICMJE Style Harshani HBC, Balasuriya DK, , Kaushalya AGG, Wijendra WAS. Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. J Microbiol Infect Dis. (2024), [cited January 03, 2026]; 14(2): 67-79. doi:10.5455/JMID.2024.v14.i2.5 Harvard Style Harshani, H. B. C., Balasuriya, . D. K., , Kaushalya, . A. G. G. & Wijendra, . W. A. S. (2024) Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. J Microbiol Infect Dis, 14 (2), 67-79. doi:10.5455/JMID.2024.v14.i2.5 Turabian Style Harshani, Hewa Babarandage Chathurika, Dilshani Kavindya Balasuriya, Wickramasinghe Widana Pathirana Pragathi Nirasha Diwyanjalee, Aranwela Gamage Gaya Kaushalya, and Wijendra Acharige Somalatha Wijendra. 2024. Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. Journal of Microbiology and Infectious Diseases, 14 (2), 67-79. doi:10.5455/JMID.2024.v14.i2.5 Chicago Style Harshani, Hewa Babarandage Chathurika, Dilshani Kavindya Balasuriya, Wickramasinghe Widana Pathirana Pragathi Nirasha Diwyanjalee, Aranwela Gamage Gaya Kaushalya, and Wijendra Acharige Somalatha Wijendra. "Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka." Journal of Microbiology and Infectious Diseases 14 (2024), 67-79. doi:10.5455/JMID.2024.v14.i2.5 MLA (The Modern Language Association) Style Harshani, Hewa Babarandage Chathurika, Dilshani Kavindya Balasuriya, Wickramasinghe Widana Pathirana Pragathi Nirasha Diwyanjalee, Aranwela Gamage Gaya Kaushalya, and Wijendra Acharige Somalatha Wijendra. "Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka." Journal of Microbiology and Infectious Diseases 14.2 (2024), 67-79. Print. doi:10.5455/JMID.2024.v14.i2.5 APA (American Psychological Association) Style Harshani, H. B. C., Balasuriya, . D. K., , Kaushalya, . A. G. G. & Wijendra, . W. A. S. (2024) Assessment of Listeria monocytogenes contamination and antibiotic resistance in raw milk samples collected from Colombo district Sri Lanka. Journal of Microbiology and Infectious Diseases, 14 (2), 67-79. doi:10.5455/JMID.2024.v14.i2.5 |