| Research Article | ||
J Microbiol Infect Dis. 2023; 13(3): 118-124 J. Microbiol. Infect. Dis., (2023), Vol. 13(3): 118–124 Original Research Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samplesSayani Roy1, Jayalaxmi Wangkheimayum2, Sanchari Roy Choudhury2, Bhaskar Jyoti Das2, Pranab Behari Mazumder1 and Amitabha Bhattacharjee2*1Department of Biotechnology, Assam University, Silchar, India 2Department of Microbiology, Assam University, Silchar, India *Corresponding Author: Adrián Melero Jurado. Exòtica Veterinaris, Barcelona, Spain. Email: adrian.vetexotic [at] hotmail.com Submitted: 16/06/2023 Accepted: 16/09/2023 Published: 30/09/2023 © 2023 Journal of Microbiology and Infectious Diseases
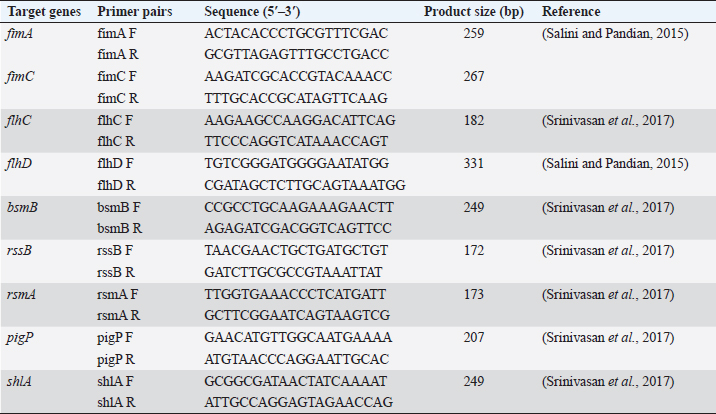
AbstractBackground: Foodborne illness is a global concern and identification of pathogens that cause foodborne disease is a public health priority. Serratia marcescens is an opportunistic pathogen responsible for food poisoning due to its ability to colonize the gastrointestinal surfaces. Serratia marcescens is also responsible for causing a wide range of extraintestinal infections such as pneumonia, urinary tract infection, and keratitis. Aim: The current study investigated the presence of antibiotic-resistant virulent S. marcescens from street food samples. Methods: Two pigmented S. marcescens were recovered from panipuri samples which were then identified using VITEK® GN cards. The virulence genes and the antibiotic resistance genes harbored by the S. marcescens isolates were investigated and further transformation assay was conducted to check the horizontal transferability of the plasmid carrying the antimicrobial resistance genes. Incompatibility typing of the transformants was also assessed and enterobacterial repetitive intergenic consensus polymerase chain reaction was performed to check the heterogeneity of the S. marcescens isolates. Results: Two S. marcescens isolates harboring virulence genes as well as antibiotic resistance genes were detected in this study and it was observed that Inc FIB type plasmid was carrying the resistance genes. Conclusion: The co-existence of both the virulence and antibiotic resistance genes in the two S. marcescens isolates warrants proper surveillance in order to prevent the spread of such pathogenic strains in the environment as well as the community through food samples. Keywords: Virulence genes, Antibiotic resistance, Serratia marcescens, Inc FIB. IntroductionMicrobial contamination of food can lead to severe health complications, leading to death in some cases. According to the World Health Organization, each year about 600 million people suffer from foodborne illnesses causing about 420,000 deaths worldwide (https://www.who.int/news-room/fact-sheets/detail/food-safety). Thus, the identification of foodborne pathogens is a global health priority. Several foodborne bacterial pathogens are involved in food contamination out of which Serratia marcescens is an emerging and less studied foodborne pathogen (Abbas and Hagazy, 2020). Serratia marcescens was considered a nonpathogenic organism until its first known involvement in a nosocomial outbreak in 1951 (Su et al., 2003). It is also considered a foodborne pathogen due to its ability to colonize a wide variety of gastrointestinal surfaces. Once inside the host body, it can penetrate deeper tissues and cause a wide variety of infections such as diarrhea, urinary tract infection, pneumonia, and osteomyelitis (Van Houdt et al., 2007). These pathogens when acquiring the antibiotic resistance determinants further exaggerate the pathogenicity of these pathogens as antibiotic resistance in them leads to serious repercussions for the treatment of any infection. The street foods have a high risk of microbial contamination due to the poor hygiene practice of the street vendors, the use of ungloved hands of vendors to serve food, and most importantly the location of their mobile food vending units as the vending units are usually located near high traffic areas exposing the food to dirt and dust. Food serves as the perfect medium for the proliferation of bacteria and a vehicle for bacterial transmission to consumers and ultimately to the community. Thus, the presence of even a very few number of enteric bacteria with virulence attributes in food poses a public health threat as such pathogens can easily proliferate and transmit to the gastrointestinal tract of consumers through food. The current study identifies two virulent S. marcescens from food samples which were co-harbouring resistance genes, implicating a possible threat to the health of street food consumers. Materials and MethodsBacterial isolatesTwo pigmented S. marcescens isolates were recovered from boiled mashed potato used in serving panipuris (fried puffy balls; a famous street food of India) in January 2022. Thirty-two samples were collected in sterile containers from street vendors selling panipuris in mobile street vending units in and around a town located in the Southern part of Assam, India. The identification of the isolates was accomplished using a VITEK® 2 compact system (Biomerieux, France) using VITEK® GN cards. VirulotypingConventional polymerase chain reaction (PCR) assay was used to target the virulence genes of S. marcescens isolates using nine specific primers as mentioned previously (Salini and Pandian, 2015; Srinivasan et al., 2017) (Table 1). T100TM thermal cycler (Bio-Rad, US) was used to perform PCR assay. PCR was performed using the following reaction conditions; initial denaturation at 95°C for 2 minutes, 34 cycles of 95°C for 1 minute, 50°C for 30 seconds, 72°C for 1 minute, and final extension of 72°C for 5 minutes. Antimicrobial susceptibility profilingThe S. marcescens isolates were subjected to antimicrobial susceptibility testing by disc diffusion test. Antibiotic discs (HiMedia, Mumbai, India) used were ampicillin (30 μg), cefuroxime (30 μg), cefalexin (30 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cefepime (30 μg), cefoxitin (30 μg), cotrimoxazole (25 μg), amikacin (30 μg), gentamicin (10 μg), tigecycline (15 μg), tetracycline (10 μg), imipenem (10 μg), meropenem (10 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), and nalidixic acid (30 μg) and the results were interpreted as per Clinical and Laboratory Standards Institute guidelines (CLSI, 2022). Molecular detection of antimicrobial resistance genesGenes encoding aminoglycoside modifying enzymes (AMEs) and tetracycline resistance were detected by PCR. The genes encoding carbapenemase as well as extended-spectrum β-lactamases (ESBLs), quinolone and sulphonamide resistance genes were also characterized by PCR assay using the previously published primers and the reaction was run according to the previously standardized reaction conditions (Rasmussen et al., 1996; Poirel and Nordmann, 2006; Woodford et al., 2006; Naas et al., 2008; Yong et al., 2009; Dallenne et al., 2010; Paul et al., 2021). Horizontal gene transferability studyThe horizontal transferability of these resistance determinants was checked by transformation assay. Plasmids were extracted from the two isolates using the QIAprep Spin Miniprep Kit (Qiagen, Germany) according to the manufacturer’s protocol. Isolated plasmids were transformed into the recipient strain Escherichia coli DH5α by heat shock method (Li et al., 2007). The transformants were then selected on Luria Bertani agar (HiMedia, India) containing 0.5 μg/ml Gentamicin. Table 1. Primer pairs targeting the virulence genes.
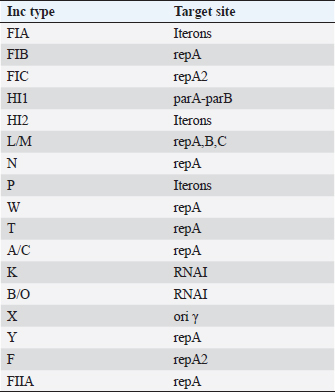
Plasmid incompatibility typingPlasmids were extracted from the transformants using the QIAprep Spin Miniprep Kit (Qiagen, Germany) according to the manufacturer’s protocol. The incompatibility typing of the plasmids was done by PCR-based replicon typing as previously described (Carattoli et al., 2005). The Inc types and their targets are mentioned in Table 2. Typing of virulent S. marcescensThe genetic similarity or heterogeneity of the two S. marcescens isolates was assessed by enterobacterial repetitive intergenic consensus (ERIC)-PCR using primers ERIC F-(ATGTAAGCTCCTGGGGATTCAC) and ERIC R-(AAGTAAGTGACTGGGGTGAGCG). PCR was performed using the following reaction conditions; initial denaturation at 95°C for 3 minutes, 30 cycles of denaturation at 95°C for 20 seconds, annealing at 46°C for 40 seconds and extension at 72°C for 3 minutes; and final extension at 72°C for 10 minutes and the banding patterns were analyzed by agarose gel electrophoresis (Versalovic et al., 1991). Table 2. Inc types and their target sites.
ResultsThe two S. marcescens (SM1 and SM2) isolates were recovered from the street food samples, i.e., boiled mashed potato used to serve panipuri. The isolate SM1 was recovered from a ready-to-eat food sample sold by a street vendor whose food vending unit was located close to a residential area and educational institutes (24°49′11.12545ʺ latitude and 92°47′43.63202ʺ longitude). This isolate harbored multiple virulence genes namely, shlA which encodes a hemolysin that is responsible for forming pores within the host membrane, bsmB which regulates bacterial adhesion, flhD is one of the genes responsible for motility (swarming and swimming) of S. marcescens and pigP responsible for producing prodigiosin (Table 3) (Figs. 1–3). The second isolate SM2 was also recovered from the boiled mashed potato sold by the ready-to-eat street food seller whose street food vending stall was located in a high pedestrian traffic area having latitude and longitude coordinates of 24°49′13.25294ʺ and 92°47′53.5979ʺ, respectively. This isolate harbored shlA and pigP virulence genes (Table 3) (Figs. 1–3). Both the street vending units were about 500 m apart and were in the vicinity of residential areas and educational institutes. Both the isolates were resistant to β-lactam (ampicillin, cephalexin, and cefuroxime), aminoglycosides (amikacin and gentamicin), and cyclin group of drugs (tigecycline and tetracycline). SM1 harbored multiple AME genes viz., ant (2ʺ)Ia, ant(3ʺ)I, aph(2ʺ)Ic, aac(3)IIc, aac(6)Ib, aac (6′)-II while isolate SM2 carried aac(6)Ib, aac(3)IIc and aph(2ʺ)Ic (Figs. 4–7). In addition, both the isolates co-carried tet(K) gene rendering tetracycline resistance (Fig. 8). Moreover, both the AMEs and tet(K) carrying plasmids were horizontally transferable and incompatibility typing of the transformants showed that the resistance genes were carried within a FIB Inc plasmid. Typing by ERIC PCR established two different clonal patterns of S. marcescens harboring both the virulence and antibiotic resistance genes.
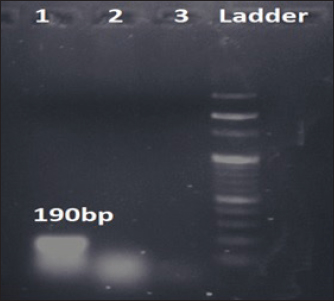
Fig. 1. Amplification of bsmB at 190 bp. Lane 1: PCR product showing amplified bsmB gene. Lane 2: negative control. Table 3. Virulence and antibiotic resistance determinants of the study isolates.
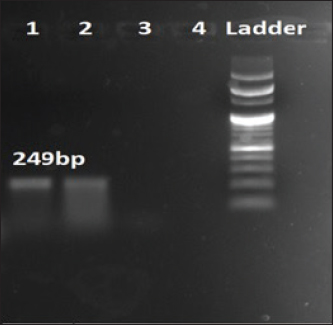
Fig. 2. Amplification of shlA at 249 bp. Lane 1, 2: PCR product showing amplified shlA gene. Lane 3: negative control.
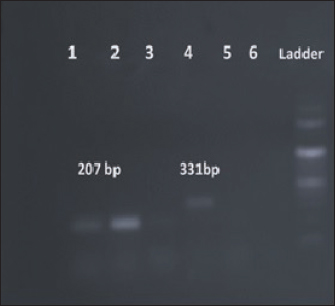
Fig. 3. Amplification of pigP at 207 bp and flhD at 331 bp. Lane 1, 2: PCR product showing amplified pigP gene. Lane 3: negative control. Lane 4: PCR product showing amplified flhD gene. Lane 5: negative control.
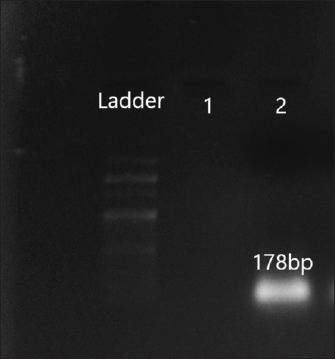
Fig. 4. Amplification of aac(6′)II at 178 bp. Lane 2: PCR product showing amplified aac (6′) II gene. Lane 1: negative control. DiscussionFoodborne illnesses account for the most common category of infectious disease according to the surveillance of centers for disease control and prevention (https://www.cdc.gov/foodsafety/outbreaks/index). Several pathogens are associated with foodborne diseases (Campos et al., 2015). Serratia marcescens, considered as one of the foodborne pathogens, is an emergent opportunistic pathogen associated with a wide spectrum of both gastrointestinal and extra-intestinal infections. Many studies reported the occurrence of S. marcescens in food samples (Melo et al., 2018; Khater et al., 2021). The pathogenesis of S. marcescens is attributed to the acquisition of virulence factors such as the production of prodigiosin, motility, production of pore-forming toxin, and resistance determinants (Srinivasan et al., 2017). These armoury of virulence factors help this pathogen to initiate and cause multiple illnesses in humans. Several virulence genes in the S. marcescens isolates have been detected in this study which is in agreement with previous studies by Aggarwal et al. (2017), Stella et al. (2021), Shanks (2013). The presence of such virulent S. marcescens isolates in street food is a threat to public health as such strains can easily disseminate to the gastrointestinal microbiota of humans through contaminated food. Once inside the host, the subsequent lysis of the host cells might help the pathogens to penetrate the tissue and spread in the host organism. Therefore, the identification of such opportunistic pathogens in food is an important global health priority.
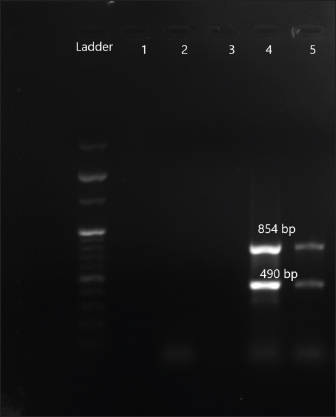
Fig. 5. Amplification of aac (3) IIc at 854 bp and aac (6)Ib at 490 bp. Lane 4,5: PCR product showing amplified aac (3) IIc gene and aac (6)Ib. Lane 3: negative control.
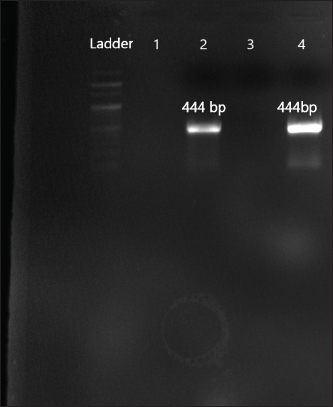
Fig. 6. Amplification of aph(2ʺ)Ic at 444 bp. Lane 2,4: PCR product showing amplified aph(2ʺ)Ic gene. Lane 1: negative control.
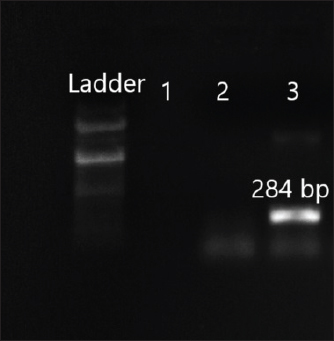
Fig. 7. Amplification of ant(3ʺ)I at 284 bp. Lane 3: PCR product showing amplified ant(3ʺ)I gene. Lane 2: negative control.
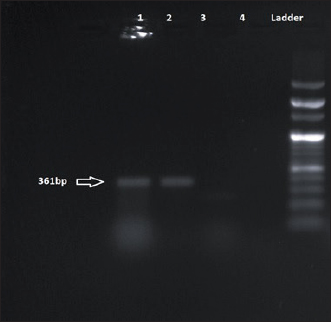
Fig. 8. Amplification of tet(K) at 361 bp. Lane 1, 2: PCR product showing amplified tet(K) gene. Lane 3: negative control. Both the study isolates were also resistant to multiple classes of antibiotics viz., β- lactams, aminoglycosides, and cycline group of drugs harboring different antibiotic resistance determinants. These findings are in concurrence with studies of de Silva et al. (2021), Moradigaravand et al. (2016). Such multiple drug-resistant Serratia strains pose a serious threat to mankind as such drug-resistant patterns would limit the treatment options available for treating life-threatening Serratia infections. Interestingly, this study has also detected an Inc FIB type plasmid which would act as a vehicle for dissemination of such multidrug resistance (MDR) genes. The concurrence of both the virulence genes and antibiotic resistance genes in our study isolates favor the increased survival of such pathogens as antibiotic resistance will limit the treatment option which may lead to the emergence of untreatable illnesses. The current study is a pilot study to investigate different enteric pathogens present within ready-to-eat food samples from this geographical part of the world. Two isolates of S. marcescens resistant to multiple antibiotics as well as harbouring virulence genes were recovered during this study. The multidrug-resistant phenotype of the study isolates and isolation sources as ready-to-eat food samples are unusual. As the presence of such phenotypes is less frequent in food samples, this implies a potential threat of the emergence of an unusual pathogen that may cause enteric diseases and other associated illnesses. Hence, reporting these two virulent MDR isolates is of public health importance to contain their spread. ConclusionIn the current study, antibiotic-resistant virulent S. marcescens was detected in the food sample. The presence of this less frequently isolated pathogen from food samples is of immense importance. The study warrants the necessity of local epidemiological investigations to understand the expansion of this pathogen to the environment and community through food samples further minimizing the threat to public health. AcknowledgmentAuthors acknowledge the infrastructural facilities provided by Advanced Institutional Biotech Hub, Assam University, Silchar, India. Authors’ contributionsSR: Experimental work, formal analysis, and writing draft; JW: Formal analysis; SRC: Sample collection and formal analysis; BJD: Formal analysis; PBM: Manuscript editing and proofreading; AB: Study design, Conceptualization, and Supervision. ReferencesAbbas, H.A. and Hegazy, W.A. 2020. Repurposing anti-diabetic drug “Sitagliptin” as a novel virulence attenuating agent in Serratia marcescens. PLoS One 15(4), e0231625. Aggarwal, C., Paul, S., Tripathi, V., Paul, B. and Khan, M.A. 2017. Characterization of putative virulence factors of Serratia marcescens strain SEN for pathogenesis in Spodoptera litura. J. Invertebr. Pathol. 143, 115–123. Campos, J., Gil, J., Mourão, J., Peixe, L. and Antunes, P. 2015. Ready-to-eat street-vended food as a potential vehicle of bacterial pathogens and antimicrobial resistance: an exploratory study in Porto region, Portugal. Int. J. Food. Microbiol. 206,1–6. Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K.L. and Threlfall, E.J. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 63(3), 219–228. CLSI. (2022). Performance standards for antimicrobial susceptibility testing 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. Dallenne, C., Da Costa, A., Decré, D., Favier, C. and Arlet, G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65(3), 490–495. da Silva, K.E., Rossato, L., Jorge, S., de Oliveira, N.R., Kremer, F.S., Campos, V.F., da Silva Pinto, L., Dellagostin, O.A. and Simionatto, S. 2021. Three challenging cases of infections by multidrug-resistant Serratia marcescens in patients admitted to intensive care units. Braz. J. Microbiol. 52(3), 1341–1345. Khater, D.F., Lela, R.A., El-Diasty, M., Moustafa, S.A. and Wareth, G. 2021. Detection of harmful foodborne pathogens in food samples at the points of sale by MALDT-TOF MS in Egypt. BMC. Res. Notes. 14(1), 1–6. Li, S., Meadow Anderson, L., Yang, J.M., Lin, L. and Yang, H. 2007. DNA transformation via local heat shock. Appl. Phys. Lett. 91(1), 1–3. Melo, D.H., Souza, B.V., Nascimento, J.S., Amorim, A.M., Medeiros, L.M. and Mattoso, J.M. 2018. A reddish problem: antibiotic-resistant Serratia marcescens in dairy food commercialized in Rio de Janeiro. Int. J. Food. Microbiol. 25(2), 880–883. Moradigaravand, D., Boinett, C.J., Martin, V., Peacock, S.J. and Parkhill, J. 2016. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome. Res. 26(8), 1101–1109. Naas, T., Cuzon, G., Villegas, M.V., Lartigue, M.F., Quinn, J.P. and Nordmann, P. 2008. Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob. Agents. Chemother. 52(4), 1257–1263. Paul, D., Mazumder, N.B., Wangkheimayum, J. and Bhattacharjee, A. 2021. Report of a carbapenemase gene blaIMP-4 in multi-drug resistant Escherichia coli from sewage water: a threat on clinical-environmental interphase. Indian. J. Med. Microbiol. 39(4), 556–557. Poirel, L. and Nordmann, P. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene bla OXA-58 in Acinetobacter baumannii. Antimicrob. Agents. Chemother. 50(4), 1442–1448. Rasmussen, B.A., Bush, K., Keeney, D., Yang, Y., Hare, R., O’Gara, C. and Medeiros, A.A. 1996. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents. Chemother. 40(9), 2080–2086. Salini, R. and Pandian, S.K. 2015. Interference of quorum sensing in urinary pathogen Serratia marcescens by Anethum graveolens. Pathog. Dis. 73(6), ftv038. Shanks, R.M., Lahr, R.M., Stella, N.A., Arena, K.E., Brothers, K.M., Kwak, D.H., Liu, X. and Kalivoda, E.J. 2013. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PloS One 8(3), e57634. Srinivasan, R., Mohankumar, R., Kannappan, A., Karthick Raja, V., Archunan, G., Karutha Pandian, S., Ruckmani, K. and Veera Ravi, A. 2017. Exploring the anti-quorum sensing and antibiofilm efficacy of phytol against Serratia marcescens associated acute pyelonephritis infection in Wistar rats. Front. Cell. Infect. Microbiol. 7, 498. Stella, N.A., Brothers, K.M. and Shanks, R.M. 2021. Differential susceptibility of airway and ocular surface cell lines to FlhDC-mediated virulence factors PhlA and ShlA from Serratia marcescens. J. Med. Microbiol. 70(2), 001292. Su, L.H., Ou, J.T., Leu, H.S., Chiang, P.C., Chiu, Y.P., Chia, J.H., Kuo, A.J., Chiu, C.H., Chu, C., Wu, T.L. and Sun, C.F. 2003. Extended epidemic of nosocomial urinary tract infections caused by Serratia marcescens. J. Clin. Microbiol. 41(10), 4726–4732. Van Houdt, R., Givskov, M. and Michiels, C.W. 2007. Quorum sensing in Serratia. FEMS. Microbiol. Rev. 31(4), 407–424. Versalovic, J., Koeuth, T. and Lupski, R. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic. Acids. Res. 19(24), 6823–6831. Woodford, N., Ellington, M.J., Coelho, J.M., Turton, J.F., Ward, M.E., Brown, S., Amyes, S.G. and Livermore, D.M. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents. 27(4), 351–353. Yong, D., Toleman, M.A., Giske, C.G., Cho, H.S., Sundman, K., Lee, K. and Walsh, T.R. 2009. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents. Chemother. 53(12), 5046–5054. | ||
| How to Cite this Article |
| Pubmed Style Roy S, Wangkheimayum J, Choudhury SR, Das BJ, Mazumder PB, Bhattacharjee A. Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. J Microbiol Infect Dis. 2023; 13(3): 118-124. doi:10.5455/JMID.2023.v13.i3.3 Web Style Roy S, Wangkheimayum J, Choudhury SR, Das BJ, Mazumder PB, Bhattacharjee A. Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. https://www.jmidonline.org/?mno=302657418 [Access: May 08, 2024]. doi:10.5455/JMID.2023.v13.i3.3 AMA (American Medical Association) Style Roy S, Wangkheimayum J, Choudhury SR, Das BJ, Mazumder PB, Bhattacharjee A. Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. J Microbiol Infect Dis. 2023; 13(3): 118-124. doi:10.5455/JMID.2023.v13.i3.3 Vancouver/ICMJE Style Roy S, Wangkheimayum J, Choudhury SR, Das BJ, Mazumder PB, Bhattacharjee A. Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. J Microbiol Infect Dis. (2023), [cited May 08, 2024]; 13(3): 118-124. doi:10.5455/JMID.2023.v13.i3.3 Harvard Style Roy, S., Wangkheimayum, . J., Choudhury, . S. R., Das, . B. J., Mazumder, . P. B. & Bhattacharjee, . A. (2023) Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. J Microbiol Infect Dis, 13 (3), 118-124. doi:10.5455/JMID.2023.v13.i3.3 Turabian Style Roy, Sayani, Jayalaxmi Wangkheimayum, Sanchari Roy Choudhury, Bhaskar Jyoti Das, Pranab Behari Mazumder, and Amitabha Bhattacharjee. 2023. Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. Journal of Microbiology and Infectious Diseases, 13 (3), 118-124. doi:10.5455/JMID.2023.v13.i3.3 Chicago Style Roy, Sayani, Jayalaxmi Wangkheimayum, Sanchari Roy Choudhury, Bhaskar Jyoti Das, Pranab Behari Mazumder, and Amitabha Bhattacharjee. "Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples." Journal of Microbiology and Infectious Diseases 13 (2023), 118-124. doi:10.5455/JMID.2023.v13.i3.3 MLA (The Modern Language Association) Style Roy, Sayani, Jayalaxmi Wangkheimayum, Sanchari Roy Choudhury, Bhaskar Jyoti Das, Pranab Behari Mazumder, and Amitabha Bhattacharjee. "Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples." Journal of Microbiology and Infectious Diseases 13.3 (2023), 118-124. Print. doi:10.5455/JMID.2023.v13.i3.3 APA (American Psychological Association) Style Roy, S., Wangkheimayum, . J., Choudhury, . S. R., Das, . B. J., Mazumder, . P. B. & Bhattacharjee, . A. (2023) Occurrence of virulent Serratia marcescens with co-existing antibiotic resistance determinants in ready-to-eat food samples. Journal of Microbiology and Infectious Diseases, 13 (3), 118-124. doi:10.5455/JMID.2023.v13.i3.3 |