| Research Article | ||
J Microbiol Infect Dis. 2024; 14(3): 109-119 J. Microbiol. Infect. Dis., (2024), Vol. 14(3): 109–119 Review Article Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from IndiaRajkumari Elizabeth1, Nikita Karmakar1, Jayalaxmi Wangkheimayum1, Bhaskar Jyoti Das1, Debadatta Dhar Chanda2 and Amitabha Bhattacharjee1*1Department of Microbiology, Assam University, Silchar, India 2Department of Microbiology, Silchar Medical College & Hospital, Silchar, India *Corresponding Author: Amitabha Bhattacharjee. Department of Microbiology, Assam University, Silchar, India. Email: ab0404 [at] gmail.com Submitted: 29/06/2024 Accepted: 02/09/2024, Published: 30/09/2024 © 2024 Journal of Microbiology and Infectious Diseases
ABSTRACTBackground: Colistin is regarded as the final option for treating Gram-negative bacteria that are resistant to several drugs. A common resistance mechanism for colistin in Klebsiella pneumoniae is mgrB gene inactivation. Inactivation of mgrB activates the PhoPQ which upregulates the PmrAB leading to resistance by lipopolysaccharide modification. Colistin resistance has been reported in Klebsiella pneumoniae still studies related to molecular mechanisms of this resistance are lacking. Aim: Role of mgrB in colistin resistance Methods: In this study, eight colistin-resistant clinical K. pneumoniae were selected, and performed broth microdilution was followed by a rapid polymyxin nordmann poirel test for screening of colistin resistance. PCR was performed for different types of mcr genes. MgrB was amplified and sequenced. Quantitative real-time PCR was performed for mgrB, PmrA, PmrB, PhoP, and PhoQ genes. Furthermore, clonal analysis by enterobacterial repetitive intergenic consensus PCR was done. Results: Colistin minimum inhibitory concentration of 4 μg/ml has been shown by the isolates. No mcr gene was detected. Positive amplification for the mgrB gene was found in all the isolates. Sequence analysis of the mgrB gene found no mutation however, higher expression in the mgrB, PmrA, PmrB, PhoP, and PhoQ genes of all the eight colistin-resistant isolates could be observed when the isolates were exposed to colistin and Mg2+ pressure. Clonal analysis exhibited seven different haplotypes of K. pneumoniae. Conclusion: This study provides firsthand knowledge about molecular mechanisms of colistin resistance in K. pneumoniae in this geographical area. Enhancement in mgrB expression in the presence of colistin and Mg2+ pressure regardless of any mutation has a remarkable impact in the context of colistin resistance mechanisms in clinical K. pneumoniae. Keywords: Colistin resistance, Klebsiella pneumoniae, mgrB, PhoPQ, PmrAB. IntroductionKlebsiella pneumoniae is one of the Gram-negative pathogens that are frequently responsible for various nosocomial infections. The emergence of multidrug-resistant K. pneumoniae and infections caused by them is now a global issue (Kocsis et al., 2017). Colistin is regarded as the final option for treating Gram-negative bacteria that are resistant to many drugs particularly carbapenemase-producing Enterobacteriaceae including Klebsiella pneumoniae. Reports of colistin-resistant Klebsiella pneumoniae isolates are rising worldwide (Poirel et al., 2015). Colistin resistance in Klebsiella pneumoniae is mainly due to the modification of lipopolysaccharide through cationic substitution caused by the activation of PhoPQ and PmrAB (two-component regulatory systems) as well as the mgrB gene’s inactivation (Cheong et al., 2020). MgrB is a small (47 amino acids) transmembrane protein that negatively regulates the PhoPQ signaling system (Poirel et al., 2015). Inactivation of mgrB gene activates the PhoPQ system which directly upregulates the PmrAB system leading to colistin resistance by modification of the lipopolysaccharide (Cannatelli et al., 2014). The lipid A component of the outer membrane lipopolysaccharide is substituted with phosphoethanolamine and 4-amino-4-deoxy-L-arabinose (L-Ara4N) interceded by the PhoPQ and PmrAB system (Wright et al., 2015). This alterations neutralizes the lipopolysaccharide’s negative charge thereby reducing the binding affinity of the colistin to lipopolysaccharide surface and thus leading to colistin resistance (Andrade et al., 2020). It was confirmed that external signals such as low Mg2+ and cationic antimicrobial peptides can activate systems responsible for lipopolysaccharide modification leading to colistin resistance in organisms like Escherichia coli and Salmonella (Richards et al., 2012). Here, we investigated whether PhoPQ, PmrAB, and mgrB react to the same physiological stimuli in Klebsiella pneumoniae to cause colistin resistance. Moreover, different types of plasmid-mediated colistin resistance mcr genes which are horizontally transferable have been reported in Enterobacteriaceae (Haeili et al., 2017). India is known for its extensive use of colistin in both humans and animals, particularly in tertiary referral hospitals (Partridge et al., 2018). There have been reports of colistin resistance in Klebsiella pneumoniae with clinical origin (Aggarwal et al., 2018). However, studies related to the underlying molecular mechanisms assisting this resistance trait are rare in this region. Therefore, it would be of tremendous interest to investigate the main mechanism triggering resistance to colistin in clinical Klebsiella pneumoniae of this geographical area and how the transcriptional regulatory systems respond to external stimuli thereby mediating resistance to this antibiotic. In the current study, we demonstrate that the expression of the transcriptional regulator mgrB in the colistin-resistant clinical Klebsiella pneumoniae isolates is enhanced upon colistin and Mg2+ exposure. Materials and MethodsBacterial isolatesIn this study, we have selected eight potentially screened colistin-resistant Klebsiella pneumoniae. The isolates were of clinical origin and were acquired from regular clinical samples processed for culture and sensitivity testing at the Department of Microbiology, Silchar Medical College and Hospital (Supplementary Table S1). The VITEK® 2 compact system (Biomeriux) was used to identify the organisms, and 16S ribosomal RNA gene sequencing was used to confirm the identification. Screening for colistin resistanceThe broth microdilution method which is a standard method for polymyxin susceptibility testing (as recommended by EUCAST guidelines 2019) was carried out using cation-adjusted Mueller-Hinton broth (Hi-Media, Mumbai, India) to screen the isolates for colistin resistance. Colistin {colistin sodium methanesulphonate (Hi-Media, Mumbai, India)} with a concentration ranging from 2 to 4 μg/ml was used for the screening and as a quality control, Escherichia coli ATCC 25922 was utilised. The outcomes were interpreted in accordance with the 2019 EUCAST criteria. Furthermore, rapid polymyxin nordmann poirel (NP) testing was used to confirm the colistin resistance of the screened positive isolates (Nordmann et al., 2016). Detection of plasmid-mediated colistin resistance mcr genesPolymerase chain reaction was used to identify the plasmid-mediated colistin resistance genes mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, mcr-6, mcr-7, mcr-8.2, mcr-8.4, mcr-9, and mcr-10 (PCR). Specific oligonucleotide primers were utilized for the corresponding genes (Supplementary Table S2). The predetermined reaction conditions were used (Elizabeth et al., 2021). PCR and sequencing of mgrB geneThe PCR assay was performed simultaneously for the amplification of the mgrB gene by utilizing certain oligonucleotide primers for the gene (Supplementary Table S2). The PCR reaction was conducted at 94°C for 3 minutes, then in 35 cycles at 94°C for 40 seconds, 51°C for 30 seconds, 72°C for 50 seconds, and finally 72°C for 7 minutes. The amplified DNA fragments were purified with the HiPurATM PCR product purification kit (Hi-Media, Mumbai, India). Sequencing of the product was outsourced by Lifetech Service Laboratory, Invitrogen Bioservices India Pvt.Ltd., India. Analysis of the sequence was done using the BLAST suite program of NCBI (http://blast.ncbi.nlm.nih.gov). Transcriptional analysis of mgrBThe expression of the mgrB gene was assessed using quantitative real-time PCR. For the analysis, oligonucleotide primers that were unique to the gene were used (Supplementary Table S2). Eight colistin-resistant were selected and one colistin-susceptible isolate [S1, minimum inhibitory concentration (MIC) against colistin; 0.5 μg/ml] was used as a reference strain. All the isolates were inoculated in 5 ml Luria–Bertani broth (Hi-Media) with colistin pressure (0.5 μg/ml of colistin), with Mg2+ (10 µM Magnesium sulfate heptahydrate, Hi-Media, Mumbai, India), with both colistin and Mg2+ and without either of them, 12–16 hours of incubation at 37°C in a shaker incubator (160 rpm). Utilizing a Qiagen RNeasy mini kit per the manufacturer’s instructions, total messenger RNA (mRNA) was extracted from mid-log bacterial cultures (OD value 0.4–0.5 at 600 nm). Using the Quantiscript Reverse Transcription kit from Qiagen, the mRNA was reverse-transcribed into complementary DNA. Using the Step One Plus real-time detection equipment from Applied Biosystem and Power Sybr Green Master Mix, quantitative real-time PCR was performed. Using the threshold cycle (ΔΔCt) approach, the fold-changes in the targeted genes’ expression under various settings were computed. The genes’ levels of transcription were quantified in triplicates and normalized against those obtained with the colistin-susceptible isolate (S1). Statistical analysis for all the genes was carried out using Student’s t-test to check the significance of the change in the expression (significant when p-value< 0.05). Transcriptional analysis of PmrAB and PhoPQBy using quantitative real-time PCR, the expression of the PmrA, PmrB, PhoP, and PhoQ genes was assessed following the same procedures as mentioned above. For each gene, specific oligonucleotide primers were utilized (Supplementary Table S2). The isolates were grown in 5 ml Luria–Bertani broth with colistin pressure, with Mg2+ and with both colistin and Mg2+ pressure and without either of them, and 12–16 hours were spent incubating at 37°C in a shaker incubator. Isolation of mRNA and cDNA was done by following the manufacturer’s instructions and quantitative real-time PCR was carried out. The threshold cycle (ΔΔCt) approach was used to determine the fold-changes in the expression and the transcription levels were normalized with that of the susceptible isolate (S1). The expression levels were quantified in triplicates for each gene under each condition. Typing of the isolates by enterobacterial repetitive intergenic consensus (ERIC) PCRTyping of the colistin-resistant Klebsiella pneumoniae isolates was performed by ERIC PCR as previously described (Versalovic et al., 1991). The reaction was conducted at 95°C for 3 minutes, then in 30 cycles for 20 seconds at 95°C, 40 seconds at 52°C, 3 minutes at 72°C, and 10 minutes at 72°C as a final extension. The result of the ERIC PCR was interpreted according to the different banding patterns and molecular weights of the isolates and a supporting dendrogram was plotted by using NTSYS software. Ethical approvalThe study was approved by Institutional Ethics Committee vide no.3 agenda no. 4, dated 9th April, 2018. ResultsBroth microdilution result showed the eight selected colistin-resistant K. pneumoniae isolates have colistin MIC of 4 μg/ml and the results of the isolates’ rapid polymyxin NP test confirmed that all the screened positive isolates were resistant to colistin. All eight selected colistin-resistant K. pneumoniae isolates showed positive amplification for the mgrB gene. The isolates were found to be devoid of any type of plasmid-mediated colistin resistance mcr gene showing that the resistance could be chromosomally mediated. The most prevalent method of colistin resistance in K. pneumoniae is due to mgrB gene inactivation through various mutations (Cannatelli et al., 2014). Therefore, the mgrB gene of all our colistin-resistant K. pneumoniae isolates was sequenced to study their genetic environment. Sequence analysis revealed no nucleotide substitutions or mutations within the gene versus those found in the mgrB gene sequences of K. pneumoniae reference strains MGH78578 (GenBank accession no. NC_009648.1) and NTUH-K2044 (GenBank accession no. NC_012731.1). Analysis of the mgrB by quantitative real-time PCR was performed to detect the effect of their expression without any nucleotide alterations and also their expression under different external stimuli. The expression of the mgrB gene in the isolates that were given either colistin or Mg2+ stress was found to be higher than the expression of isolates without any stress and the expression was found to be highest for the isolates with both colistin and Mg2+ exposure and in one isolate we could found an increase in expression level upto six-fold (Fig. 1). Enhancement in the expression of PmrA was observed in the colistin-resistant isolates upto three-fold when the isolates were given colistin and Mg2+ stress than the isolates that were grown without any stress (Fig. 2). In the case of PmrB, more expression was seen when the isolates were administered with both colistin and Mg2+ stress when compared to isolates grown under no pressure (Fig. 3). Either colistin or Mg2+ pressure enhanced the PhoP expression however, in four isolates we could observe the highest expressional pattern upto two fold when compared with the expression of the isolates grown without any type of pressure (Supplementary Fig. S1). Similar expression patterns were seen for the phoQ gene; however, in one isolate, the expression was enhanced upto two-fold was observed when the isolate was given only Mg2+ stress (Supplementary Fig. S2). The differences in the expression of the genes were found to be significant (p-value is less than 0.05). Overall, transcriptional expressional studies showed that external stimuli such as colistin and Mg2+ pressure enhanced the expressional pattern of mgrB, PmrA and PmrB, PhoP, and PhoQ with the colistin-resistant isolates (Table 1). Clonal analysis of the eight colistin-resistant isolates with the mgrB gene revealed seven different haplotypes of Klebsiella pneumoniae and their relatedness was depicted in the dendrogram (Supplementary Fig. S3). DiscussionClinical K. pneumoniae isolates have been found to be colistin-resistant in different parts of the world (Cheong et al., 2020). Colistin is widely utilised in human beings in India, primarily in tertiary hospitals, therefore resistance towards this antibiotic is increasing (Partridge et al., 2018). Abdul Ghafur in 2014, reported colistin-resistant Gram-negative bacteria among patients with bacteremia highlighting the emergence of colistin resistance due to the increasing and inappropriate colistin usage in various tertiary hospitals in India (Ghafur et al., 2014). Another study from India also reported clinical K. pneumoniae showing resistance to colistin (Pragasam et al., 2017). Colistin resistance in clinical K. pneumoniae isolates has been reported however, information related to the molecular mechanisms mediating colistin resistance is rare in this part of the world. Therefore, our study centered on the systems regulating colistin resistance in clinical K. pneumoniae of this region. In the current study, the tested isolates (Klebsiella pneumoniae) were found to be showing colistin MIC of 4 μg/ml which is above the breakpoint, i.e., 2 μg/ml according to the EUCAST recommendations (EUCAST version 9.0, 2019; (http://www.eucast.org) which is in agreement with other studies (Poirel et al., 2015; Hamel et al., 2020).
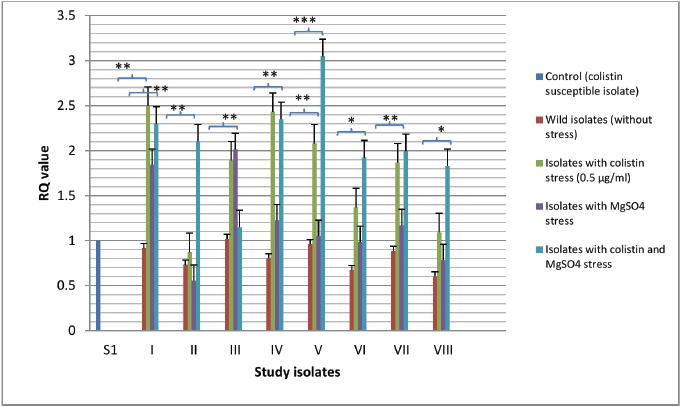
Fig. 1. Expressional pattern of mgrB. The relative quantity (RQ) values of the expression of mgr B gene in the isolates are plotted in the Y-axis. S1 is the control strain (RQ=1). Asterisks designate significance (***p < 0.001, **p < 0.01).
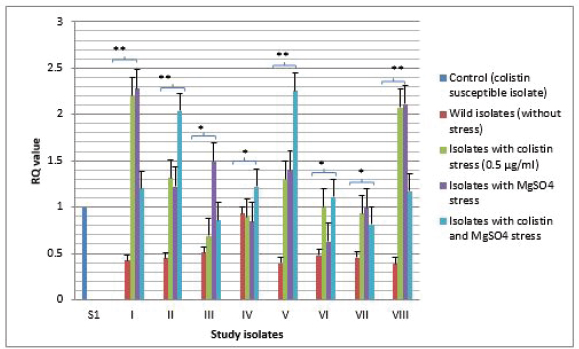
Fig. 2. Expressional pattern of PmrA. The relative quantity (RQ) values of the expression of PmrA gene in the isolates are plotted in the Y-axis. S1 is the control strain (RQ=1). Asterisks designate significance (***p < 0.001, **p < 0.01, *p < 0.05)
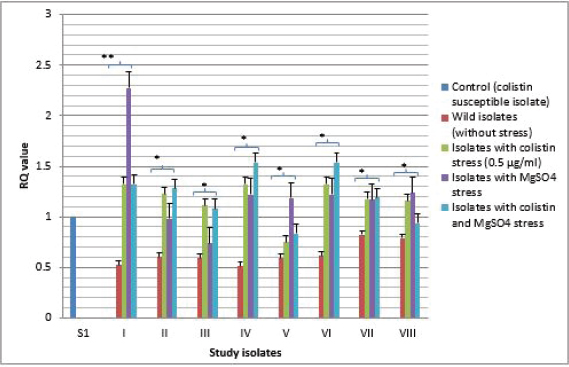
Fig. 3. Expressional pattern of PmrB. The relative quantity (RQ) values of the expression of PmrB gene in the isolates are plotted in the Y-axis. S1 is the control strain (RQ=1). Asterisks designate significance (**p < 0.01, *p < 0.05).
Supplementary Fig. S1. Expressional pattern of PhoP. The relative quantity (RQ) values of the expression of PhoP gene in the isolates are plotted in the Y-axis. S1 is the control strain (RQ=1). Asterisks designate significance (**p < 0.01, *p < 0.05). The present study is retrospective in nature and all the study isolates had clinical origin and were associated with urinary tract infection and wound infection. Due to the retrospective nature of the study, the patients could not be traced to prior colistin administration before being admitted to the tertiary care hospital. In the present study, plasmid-encoded mcr genes were not detected in any of the tested colistin-resistant K. pneumoniae of our study. This demonstrates that the resistance may be mediated by some other chromosomally mediated mechanisms. Various studies have shown that insertional inactivations within the mgrB gene were involved in resistance to colistin in K. pneumoniae (Cannatelli et al., 2013; Jayol et al., 2014; Poirel et al., 2015). Colistin resistance mediated by mutated mgrB was reported by Giordano et al. (2018). In Moscow, colistin-resistant K. pneumoniae isolates with inactivated mgrB gene by insertion element MITEKpn1 were found (Shamina et al., 2020). On the contrary, in our study, mgrB was uniformly present in our tested colistin-resistant isolates and we could not find any insertion sequences or nucleotide substitutions in mgrB in the colistin-resistant K. pneumoniae isolates. In South India, Kumar et al., (2018) described for the first time that insertional inactivation of mgrB by insertion of ISKpn14 at various locations results in colistin resistance in K. pneumoniae. Our study is in contrast to their study as we could not find any mutation or any insertion element within the mgrB gene; however, expression of mgrB was observed which may be accountable for causing colistin resistance in our K. pneumoniae isolates of this north-eastern region which is a very unusual and interesting finding. This finding is also in contrast to the study conducted by Poirel et al. (2015) who stated that the inactivation of mgrB mediates acquired colistin resistance in K. pneumoniae (Poirel et al., 2015). However, a report from Hungary agrees with our finding as they have detected a novel mgrB variant without any mutations in colistin-resistant K. pneumoniae (Kocsis et al., 2017). Enhance in the expression of the targeted genes, i.e., mgrB, PmrA, PmrB, PhoP, and PhoQ was observed in the colistin-resistant isolates in the presence of a sub-inhibitory concentration of colistin and Mg2+. This finding suggests that in colistin-resistant Klebsiella pneumoniae, exposure to colistin and Mg2+ may alter the expression patterns of mgrB and two-component regulatory systems. This can be connected to other studies that outlined the turning on of the PhoPQ system in the presence of low concentrations of Mg2+ and colistin which further activates PmrAB, which stimulates the modifications of the lipopolysaccharide mediating colistin resistance (Garcia Vescovi et al., 1996; Bader et al., 2005; Richards et al., 2012; Kang et al., 2019). Although we could not find any nucleotide alterations within the mgrB gene like in other studies, we were able to determine that the expression of mgrB is increased when there is a sub-inhibitory concentration of colistin and Mg2+ in the colistin-resistant K. pneumoniae isolates. Data from this investigation showed that the expression of the transcriptional regulator mgrB is enhanced irrespective of their mutational pattern in the colistin-resistant K. pneumoniae. Therefore, our findings constitute a striking influence in the scenario of resistance mechanisms to colistin in clinical K. pneumoniae of this region.
Supplementary Fig. S2. Expressional pattern of PhoQ. The relative quantity (RQ) values of the expression of PhoQ gene in the isolates are plotted in the Y-axis. S1 is the control strain (RQ=1). Asterisks designate significance (**p < 0.01, *p < 0.05).
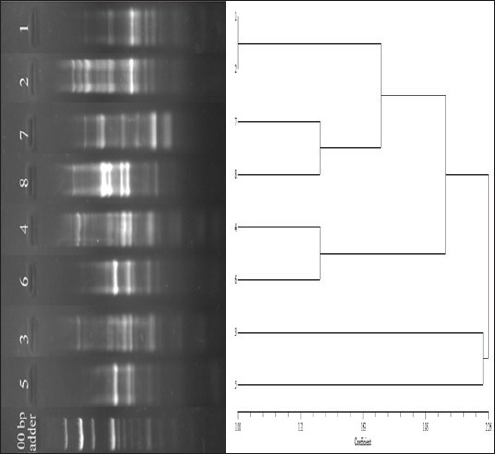
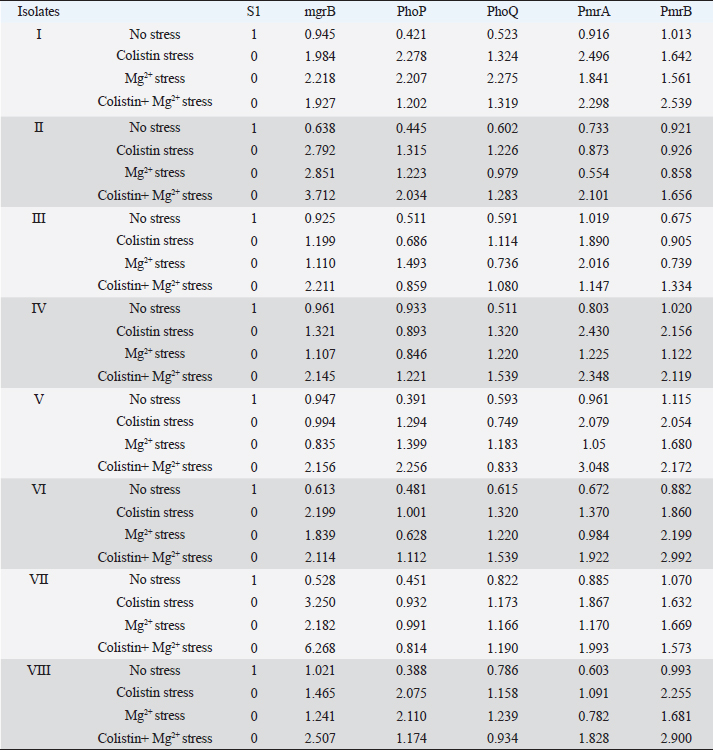
Supplementary Fig. S3. Dendrogram showing clonal diversity of the studied eight K. pneumoniae isolates by ERIC-PCR. 7 different haplotypes of K. pneumoniae was identified. Table 1. Representing the relative quantity (RQ) of the expression of the genes in different conditions shown by the isolates.
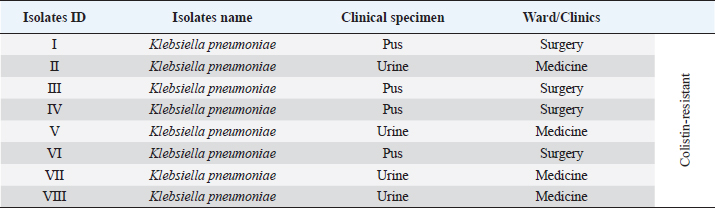
Supplementary Table S1. Clinical details of the isolates.
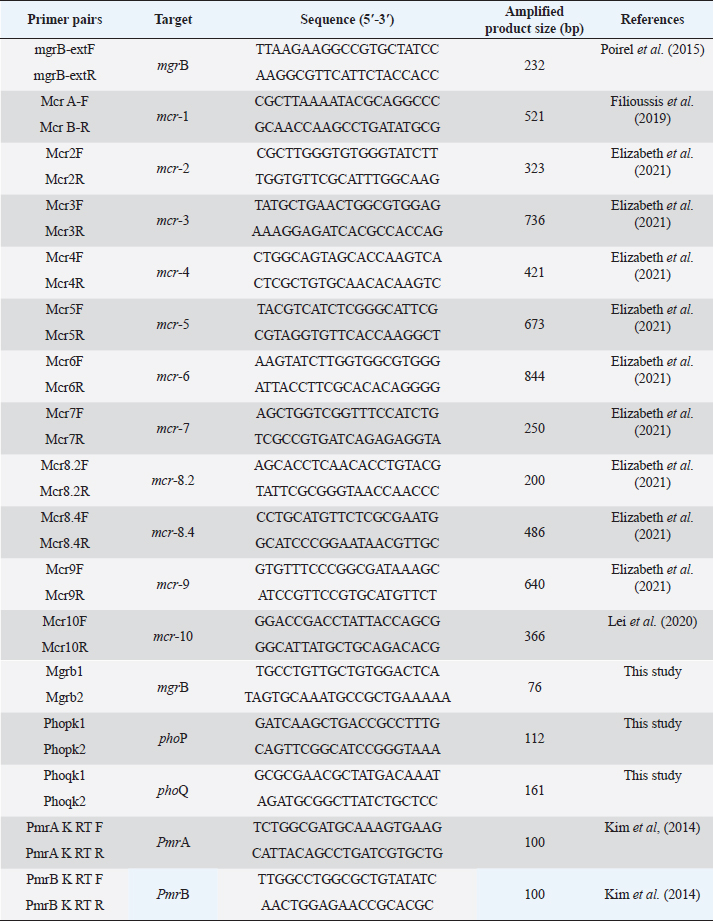
Supplementary Table S2. Oligonucleotides used in this study.
ConclusionThis study has shown that insertional inactivation in the transcriptional regulatory mgrB gene is not the only consequence influencing colistin resistance in K. pneumoniae enhancement in the expression of mgrB in the presence of colistin and Mg2+ pressure regardless of any mutation or nucleotide alterations within the gene provides a remarkable impact in the situation of colistin resistance mechanisms in clinical K. pneumoniae. This is the first study on the molecular causes of colistin resistance in isolates of K. pneumoniae in this geographical area. AcknowledgmentsThe authors would like to acknowledge the Head, Department of Microbiology, Assam University, and Assam University Advance level Biotech Hub, for providing the infrastructural support. Conflicts of interestThe authors declare that there is no conflict of interest. FundingNil. Authors’ contributionsRE: design and performed the experimental work, literature search, data collection, analysis and prepared the manuscript. NK: participated in experiment and analysis. JW: participated in experiment. BJD: participated in experiment. DD: participated in experiment designing and manuscript correction. AB: supervised the research work and participated in designing the study and checking and finalizing the manuscript. All authors read and approved the final manuscript. Data availabilityAll data are provided in the manuscript. ReferencesAggarwal, R., Rastogi, N., Mathur, P., Soni, K.D., Kumar, S., Gupta, A. and Sagar, S. 2018. Colistin-resistant Klebsiella pneumoniae in surgical polytrauma intensive care unit of level-1 trauma center: first case series from trauma patients in India. Indian J. Crit. Care Med. 22(2), 103–106. Andrade, F.F., Silva, D., Rodrigues, A. and Pina-Vaz, C. 2020. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 8, 1716. Bader, M.W., Sanowar, S., Daley, M.E., Schneider, A.R., Cho, U., Xu, W., Klevit, R.E., Le Moual, H. and Miller, S.I. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. Cannatelli, A., D’Andrea, M.M., Giani, T., Pilato, V.D., Arena, F., Ambretti, S., Gaibani, P. and Rossolini, G.M. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-Type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57(11), 5521–5526. Cannatelli, A., Giani, T., D’Andrea, M.M., Pilato, V.D., Arena, F., Conte, V., Tryfinopoulou, K., Vatopoulos, A., Rossolini, G.M. and COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. Cheong, H.S., Kim, S.Y., Seo, J., Wi, Y.M., Peck, K.R. and Ko, K.S. 2020. Colistin resistance and extensive genetic variations in PmrAB and PhoPQ in Klebsiella pneumoniae. Isolates from South Korea. Curr. Microbiol. 77, 2307–2311. Elizabeth, R., Wangkheimayum, J., Singha, K.M., Chanda, D.D. and Bhattacharjee, A. 2021. Subinhibitory concentration stress of colistin enhanced PhoPQ expression in Escherichia coli harboring mcr-1. J. Basic Microbiol. 61(11), 1–6. Filioussis, G., Kachrimanidou, M., Christodoulopoulos, G., Kyritsi, M., Hadjichristodoulou, C., Adamopoulou, M., Tzivara, A., Kritas, S.K. and Grinberg. A. 2019. Bovine mastitis caused by a multidrug-resistant, mcr-1 positive (colistin-resistant), extended-spectrum β-lactamase producing Escherichia coli clone on a Greek dairy farm. J. Dairy Sci.103, 852–857. Garcia Vescovi, E., Soncini, F.C. and Groisman, E.A. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174. Ghafur, A., Vidyalakshmi, P.R., Murali, A., Priyadarshini, K. and Thirunarayan, M.A. 2014. Emergence of pan-drug resistance amongst gram negative bacteria! The first case series from India. J. Microbiol. 4(3), 86–91. Giordano, C., Barnini, S., Tsioutis, C., Chlebowicz, M.A., Scoulica, E.V., Gikas, A., Rossen, J.W., Friedrich, A.W. and Bathoorn, E. 2018. Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of ISL3 on plasmids. Int. J. Antimicrob. Agents. 51, 260–265. Haeili, M., Javani, A., Moradi, J., Jafari, Z., Feizabadi, M. and Babaei, E. 2017. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front. Microbiol. 8, 2470. Hamel, M., Chatzipanagiotou, S., Hadjadj, L., Petinaki, E., Papagianni, S., Charalampaki, N., Tsiplakou, S., Papaioannou, V., Skarmoutsou, N., Spiliopoulou, I., Christofidou, M., Papamichalopoulos, N., Skalidis, T., Legakis, N., Fountoulis, K., Perivolioti, E., Kraniotaki, H., Bournia, M., Ioannidis, A., Baron Sophie, A. and Rolain, J.M. 2020. Inactivation of mgrB gene regulator and resistance to colistin is becoming endemic in carbapenem-resistant Klebsiella pneumoniae in Greece: a nationwide study from 2014 to 2017. Int. J. Antimicrob. Agents. 55, 105930. Jayol, A., Poirel, L., Brink, A., Villegas, M.V., Yilmaz, M. and Nordmann, P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58(8), 4762–4766. Kang, K.N., Klein, D.R., Kazi, M.I., Guerin, F., Cattoir, V., Brodbelt, J.S. and Boll, J.M. 2019. Colistin heteroresistance in Enterobacter cloacae is mediated by PmrAB-independent 4-amino-4-deoxy-L-arabinose addition to lipid A. bioRxiv; doi: http://dx.doi.org/10.1101/516872 Kim, S.Y., Choi, H.J. and Ko, K.S. 2014. Differential expression of two-component systems, pmrAB and phoPQ, with different growth phases of Klebsiella pneumoniae in the presence or absence of colistin. Curr. Microbiol. 69, 37–41. Kocsis, B., Kadar, B., Toth, A., Fullar, A. and Szabo, D. 2017. MgrB variants in colistin-susceptible and colistin-resistant Klebsiella pneumoniae ST258. J. Microbiol. Immunol. Infect. 50, 735–736. Kumar, A., Biswas, L., Omgy, N., Mohan, K., Vinod, V., Sajeev, A., Nair, P., Singh, S. and Biswas, R. 2018. Colistin resistance due to insertional inactivation of the mgrB in Klebsiella pneumoniae of clinical origin: first report from India. Rev. Esp. Quimioter. 31, 406–410. Lei, C.W., Zhang, Y., Wang, Y.T. and Wang, H.N. 2020. Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from Enterobacter roggenkampii of chicken origin in China. Antimicrob. Agents Chemother. 64, 01191–01120. Nordmann, P., Jayol, A. and Poirel, L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg. Infect. Dis. 22, 6. Partridge, S., Pilato, D.V., Doi, Y., Feldgarden, M., Haft, D. and Klimke, W. 2018. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. Poirel, L., Jayol, A., Bontron, S., Villegas, M.V., Ozdamar, M., Turkoglu, S. and Nordmann, P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. Pragasam, A.K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L.E., Inbanathan, F.Y., George, B. and Verghese, S. 2017. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India-a first report. Front. Microbiol. 7, 2135. Richards, S.M., Strandberg, K.L., Conroy, M. and Gunn, J.S. 2012. Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front. Cell. Infect. Microbiol. 2, 102. Shamina, O.V., Kryzhanovskaya, O.A., Lazareva, A.V., Alyabieva, N.M., Polikarpova, S.V., Karaseva, O.V. and Mayanskiy, N.A. 2020. Emergence of a ST307 clone carrying a novel insertion element MITEKpn1 in the mgrB gene among carbapenem-resistant Klebsiella pneumoniae from Moscow, Russia. Int. J. Antimicrob. Agents. 55, 105850. The European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. Available via http://www.eucast.org Versalovic, J., Koeuth, T. and Lupski, J.R. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. Wright, M.S., Suzuki, Y., Jones, M.B., Marshall, S.H., Rudin, S.D., van Duin, D., Kaye, K., Jacobs, M.R., Bonomo, R.A. and Adams, M.D. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. | ||
| How to Cite this Article |
| Pubmed Style Elizabeth R, Karmakar N, Wangkheimayum J, Das BJ, (chanda) DD, Bhattacharjee DA. Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. J Microbiol Infect Dis. 2024; 14(3): 109-119. doi:10.5455/JMID.2024.v14.i3.4 Web Style Elizabeth R, Karmakar N, Wangkheimayum J, Das BJ, (chanda) DD, Bhattacharjee DA. Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. https://www.jmidonline.org/?mno=207467 [Access: July 12, 2025]. doi:10.5455/JMID.2024.v14.i3.4 AMA (American Medical Association) Style Elizabeth R, Karmakar N, Wangkheimayum J, Das BJ, (chanda) DD, Bhattacharjee DA. Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. J Microbiol Infect Dis. 2024; 14(3): 109-119. doi:10.5455/JMID.2024.v14.i3.4 Vancouver/ICMJE Style Elizabeth R, Karmakar N, Wangkheimayum J, Das BJ, (chanda) DD, Bhattacharjee DA. Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. J Microbiol Infect Dis. (2024), [cited July 12, 2025]; 14(3): 109-119. doi:10.5455/JMID.2024.v14.i3.4 Harvard Style Elizabeth, R., Karmakar, . N., Wangkheimayum, . J., Das, . B. J., (chanda), . D. D. & Bhattacharjee, . D. A. (2024) Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. J Microbiol Infect Dis, 14 (3), 109-119. doi:10.5455/JMID.2024.v14.i3.4 Turabian Style Elizabeth, Rajkumari, Nikita Karmakar, Jayalaxmi Wangkheimayum, Bhaskar Jyoti Das, Dr.debadatta Dhar (chanda), and Dr. Amitabha Bhattacharjee. 2024. Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. Journal of Microbiology and Infectious Diseases, 14 (3), 109-119. doi:10.5455/JMID.2024.v14.i3.4 Chicago Style Elizabeth, Rajkumari, Nikita Karmakar, Jayalaxmi Wangkheimayum, Bhaskar Jyoti Das, Dr.debadatta Dhar (chanda), and Dr. Amitabha Bhattacharjee. "Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India." Journal of Microbiology and Infectious Diseases 14 (2024), 109-119. doi:10.5455/JMID.2024.v14.i3.4 MLA (The Modern Language Association) Style Elizabeth, Rajkumari, Nikita Karmakar, Jayalaxmi Wangkheimayum, Bhaskar Jyoti Das, Dr.debadatta Dhar (chanda), and Dr. Amitabha Bhattacharjee. "Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India." Journal of Microbiology and Infectious Diseases 14.3 (2024), 109-119. Print. doi:10.5455/JMID.2024.v14.i3.4 APA (American Psychological Association) Style Elizabeth, R., Karmakar, . N., Wangkheimayum, . J., Das, . B. J., (chanda), . D. D. & Bhattacharjee, . D. A. (2024) Expression of transcriptional regulator mgrB is enhanced in colistin-resistant Klebsiella pneumoniae: A report from India. Journal of Microbiology and Infectious Diseases, 14 (3), 109-119. doi:10.5455/JMID.2024.v14.i3.4 |