| Research Article | ||
J Microbiol Infect Dis. 2023; 13(4): 171-174 J. Microbiol. Infect. Dis., (2023), Vol. 13(4): 171–174 Original Research Molecular detection of Escherichia coli isolated from human, animal, and environmental samples targeting mdh gene: A simple one health approachUrmy Biswas, Abhi Mallick, Surojit Das*Biomedical Laboratory Science and Management, Vidyasagar University, Midnapore, India *Corresponding Author: Surojit Das. Biomedical Laboratory Science and Management, Vidyasagar University, Midnapore, India. Email: esurojitdas [at] gmail.com Submitted: 01/06/2023 Accepted: 14/12/2023 Published: 31/12/2023 © 2023 Journal of Microbiology and Infectious Diseases
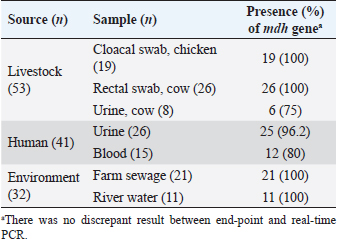
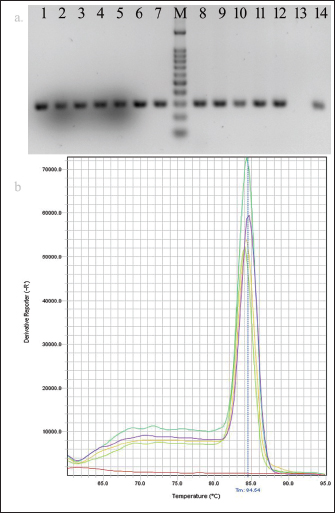
AbstractBackground: Escherichia coli cause a wide range of infections in humans and animals in hospitals and community associated with multidrug-resistance, and serves as a bioindicator of fecal contamination as well as antimicrobial resistance (AMR). Aim: Early detection of this bioindicator from humans, animals, and environment facilitates characterization with respect to AMR and bacterial forensics for effective public health and infection control measurements. Method: We investigated a simple and rapid species-specific end-point and real-time PCR assays targeting malate dehydrogenase (mdh) gene to detect E. coli isolated from livestock, human, and environmental water in Midnapore, West Bengal, India, in a timely and cost-effective approach. Results: Of 126 E. coli isolates, 53, 41, and 32 were cultured from livestock, human, and environmental water. The mdh gene was present in 120 (95.2%) isolates, including all environmental isolates. Two (3.8%) animal and four human (9.8%) isolates were discordant between culture and PCR results. The PCR assays produced results in about 90 minutes with a running cost of US$3-5 per sample. Conclusion: Both the PCR assays may be utilized in detection of E. coli depending on the laboratory infrastructure. Keywords: Escherichia coli, Bioindicator, PCR, mdh. IntroductionEscherichia coli (E. coli), which belongs to the family Enterobacterales, is of high public health concern due to worldwide increases in antimicrobial resistance (AMR) (Anjum et al., 2021). Escherichia coli is the common gut microbiota of humans and warm-blooded animals, as well as anthropogenic-mediated environmental contaminants that serve as a bioindicator of circulating AMR due to their early adaptation to antibiotic resistance genes (Anjum et al., 2021). This pathogen is frequently associated with community and hospital outbreaks leading to case fatalities (Poirel et al., 2018). The World Health Organization recognized critical priority E. coli as one health challenge (Fuga et al., 2022). Molecular techniques depreciate the traditional microbiological methods in microbial identification with respect to time and cost (Tran et al., 2015). End-point and real-time PCRs are commonly used as molecular biology tools, depending on the infrastructure and equipment facilities of the laboratory (Tarr et al., 2002; Molina et al., 2015). Here, we standardized the molecular detection of E. coli, collected from different sources targeting a housekeeping gene, malate dehydrogenase (mdh) by end-point and real-time PCRs at a low cost and in a short time for easier implementation. Material and MethodsThe prospective study was conducted in the Department of Biomedical Laboratory Science and Management at Vidyasagar University, Midnapore, West Bengal, India. Escherichia coli isolates (n=126) used in this study were collected from livestock, human, and environmental water in Midnapore district from August 2021 to March 2022. Rectal swabs of livestock were transported in Carry-Blair medium and enriched in 10 ml brain-heart infusion (BHI) broth (BD, Sparks, MD) followed by sub-cultured on eosin-methylene blue (EMB) agar. Sewage and river water samples were collected in sterile 500 ml containers and filtered through 0.45 µm pore membranes (50 ml) using filtration assembly followed by inoculation on EMB agar plates. The EMB agar-derived metallic-sheen colonies were considered for suspected E. coli isolates following confirmation by standard biochemical tests. Clinical E. coli isolates were collected from a healthcare in Midnapore, West Bengal. The isolates were preserved in BD with 15% glycerol (v/v) at −70°C. The biochemical-confirmed isolates were screened for the presence of the mdh gene by end-point and real-time PCRs using published primers (forward: 5’-GGTATGGATCGTTCCGACCT-3’; reverse: 5’-GGCAGAATGGTAACACCAGAGT-3’; amplicon size: 304 bp) with modification (Tarr et al., 2002). Quality control was assessed using the strains E. coli ATCC 25922, Klebsiella pneumoniae ATCC700603, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923. The boiled DNA templates were prepared by dissolving a single colony in 200 μl of distilled water, lysing the bacteria at 95°C for 10 minutes, and collecting the supernatant following centrifugation. The 15 µl amplification mixture was prepared for end-point or real-time PCR using 7.5 µl of 2XTaq Mix (SRL, India) or RealQ Plus 2X Master Mix Green (AMPLIQON, Denmark), 0.15 µl of 20 µM of each primer, 2 µl of template DNA, and 5.2 µl of molecular-grade water. End-point PCR was performed on a Bio-Rad T100 thermocycler (Bio-Rad, US) at 95°C for 3 minutes, followed by 30 cycles of 30 seconds denaturation at 95°C, 30 seconds annealing at 60°C, 30 seconds extension at 72°C and a final extension step at 72°C for 5 minutes. PCR amplicons were analyzed on pre-EtBr stained 2% agarose gels at 100 V for 20 minutes and visualized using the Gel-Doc system (Bio-Rad). PCR products were purified using for Sanger sequencing. Real-time PCR was carried out on a StepOnePlus system (ThermoFisher Scientific, CA, USA) with a two-step program at 95°C for 15 minutes, followed by 40 cycles of denaturation at 95°C for 20 seconds, and annealing-extension at 60°C for 60 seconds. The melt curve analysis was performed for 15 seconds at 95°C, 60 seconds at 60°C, and 15 seconds at 95°C (ramp rate of 0.3°C/second and step and hold acquisition). The StepOnePlus software ver. 2.3 was used for the analysis of real-time PCR data. A cut-off cycle threshold (Ct) value ≤35 was considered a positive result. ResultsOf 126 E. coli isolates, livestock isolates (n=53) were cultured from chicken cloacal swabs (n=19), cow rectal swabs (n=26), and urine (n=8). Human isolates (n=41) were isolated from urine (n=26) and blood (n=15) samples (Table 1). Environmental isolates (n =32) were derived from farm sewage (n=21) and Kasai river water (n=11). Amplification with the primer pair of the mdh gene showed a single band of 304 bp and a melt-curve peak at 84.5°C, indicating the specificity of end-point and real-time PCR assays, respectively (Fig. 1). Sanger sequencing results were matched with the mdh gene of E. coli by nucleotide BLAST program of the NCBI. The mdh gene was present in 120 (95.2%) isolates covering all environmental isolates (Table 1). The 51 (96.2%) animal and 37 (90.2%) human isolates showed concordance between culture and PCR (Table 1). No discrepancy was noted among the two PCR assays. The median Ct value was 23.5 (range, 16.3–29.8) for positive samples. Amplification specificities were determined by gel-electrophoresis in end-point PCR and melt-curve analysis in real-time PCR, respectively. Both the PCR assays produced results in about 90 minutes, including hands-on time. The cost of consumables and running for the assays were US$3 and US$5 per sample, respectively. Table 1. Identification of culture-confirmed E. coli isolates from different sources (n=126) targeting mdh gene by end-point and real-time PCR.
DiscussionThe commensal E. coli benefits human and animal guts, while the pathogenic one causes a wide range of infections, from diarrhea to sepsis. PCR-based approach detects the fecal contaminants like E. coli in various samples, especially food and the environment, in a rapid way. The occurrence of E. coli in hospital environments alerts for nosocomial infections and AMR (Poirel et al., 2018). Researchers reported the resistance to life-saving antibiotics, including carbapenems and colistin in human-, livestock-, and environment-derived E. coli (Barlaam et al., 2019; Das, 2023). Rapid detection of E. coli facilitates immediate health and infection control measures, including phylogrouping, pathotyping, AMR profiling, and clonal assessment (Fuga et al., 2022). The close genetic proximity of E. coli to Shigella spp. challenges the molecular discrimination of these organisms (Ivanetich et al., 2006; Omar and Barnard, 2014). Several researchers targeted different genes of E. coli by PCR, including uidA, mdh, and yaiO. Primer blast analysis of the uidA and yaiO genes showed hits with Shigella boydii, Shigella flexneri, and Shigella sonnei, while the mdh primer matched with S. flexneri only (Molina et al., 2015; Ivanetich et al., 2006). Hence, the mdh gene was selected for molecular identification of E. coli in this study. Both cost and time impact the implementation and utility of the detection method. Microbial identification of Enterobacteriaceae by biochemical analysis requires a turn-around-time of 48–72 hours with an operational cost of US$300, while the matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF) produces results in 30 minutes costing US$40 (Tran et al., 2015). Molecular methods employing 16S rRNA gene sequencing completes in 48 hours costing US$100 (Liu et al., 2021). Tarr et al. (2002) detected E. coli in about 2 hours by mdh-based end-point PCR with a running cost of US$20. In this study, we optimized the mastermix composition and thermal-cycling condition of both PCR assays to produce results in 90 minutes costing US$3-5 per sample and facilitating early downstream management of this indicator organism. Both PCR assays performed similarly in detecting the mdh gene. Six study isolates were biochemically screened as E. coli showing the absence of the mdh gene (Table 1). Discrepant isolates were checked by the VITEK2 system (bioMérieux SA, Marcy l’Etoile, France), which interpreted two of six as E. coli (data not shown). Omar and Barnard reported the absence of the mdh gene in 15% of clinical E. coli isolates (Omar and Barnard, 2014).
Fig. 1. Data analysis of PCR. (a) Agarose gel electrophoresis showing amplification of mdh gene (304 bp): samples, lanes 1–12; negative control, lane 13; positive control, lane 14 and 100 bp DNA ladder, lane M. (b) Melt curve showing a single peak at Tm of 84.5°C. ConclusionBoth the PCR assay formats targeting the mdh gene may be implemented depending on the infrastructure and equipment facilities of the laboratory to detect E. coli in different samples. Real-time approach needs minimal hands-on time over end-point PCR. AcknowledgmentsWe would like to thank the Mili Barik, project staff of Vidyasagar University for their technical support in the study. We convey thanks to Prof. Ajoy Misra, Director, Instrumental Facility at University Science Instrumentation Centre (USIC), Vidyasagar University for providing the instrumental support. Conflicts of interestAll authors report no conflicts of interest relevant to this article. Financial supportThe financial support for this work came from the University Grant Commission, India (Grant: 30-518/2020). Ethical approvalThe present study was approved by the Ethical committee of Vidyasagar University (VU/IEC-2/7-SD/19). ReferencesAnjum, M.F., Schmitt, H., Börjesson, S., Berendonk, T.U. and WAWES Network. 2021. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. 62, 152–158; doi:10.1016/j.mib.2021.09.011 Barlaam, A., Parisi, A., Spinelli, E., Caruso, M., Di Taranto, P. and Normanno, G. 2019. Global emergence of colistin-resistant Escherichia coli in food chains and associated food safety implications: a review. J. Food Protect. 82, 1440–1448; doi: 10.4315/0362-028X.JFP-19-116 Das, S. 2023. The crisis of carbapenemase-mediated carbapenem resistance across the human-animal-environmental interface in India. Infect. Dis. Now. 53, 104628; doi:10.1016/j.idnow.2022.09.023 Fuga, B., Sellera, F.P., Cerdeira, L., Esposito, F., Cardoso, B., Fontana, H., Moura, Q., Cardenas-Arias, A., Sano, E., Ribas, R.M. and Carvalho, A.C. 2022. WHO critical priority Escherichia coli as one health challenge for a post-pandemic scenario: genomic surveillance and analysis of current of trends in Brazil. Genomics Proteomics 10, 1256–1277; doi:10.1128/spectrum.01256-21 Ivanetich, K.M., Hsu, P.H., Wunderlich, K.M., Messenger, E., Walkup IV, W.G., Scott, T.M., Lukasik, J. and Davis, J. 2006. Microbial source tracking by DNA sequence analysis of the Escherichia coli malate dehydrogenase gene. J. Microbiol. Methods 67, 507–526; doi: 10.1016/j.mimet.2006.04.026 Liu, Y.X., Qin, Y., Chen, T., Lu, M., Qian, X., Guo, X. and Bai, Y. 2021. Practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 12, 315–330; doi:10.1007/s13238-020-00724-8 Molina, F., López-Acedo, E., Tabla, R., Roa, I., Gómez, A. and Rebollo, J.E. 2015. Improved detection of Escherichia coli and coliform bacteria by multiplex-PCR. BMC Biotechnol. 15, 48; doi:10.1186/s12896-015-0168-2 Omar, K.B. and Barnard, T.G. 2014. Detection of diarrhoeagenic Escherichia coli in clinical and environment water sources in South Africa using single-step 11-gene m-PCR. World J. Microbiol. Biotechnol. 30, 2663–2671; doi:10.1007/s11274-014-1690-4 Poirel, L., Madec, J.Y., Lupo, A., Schink, A.K., Kieffer, N., Nordmann, P. and Schwarz, S. 2018. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6(4), 6–4; doi:10.1128/microbiolspec.arba-0026-2017 Tarr, C.L., Large, T.M., Moeller, C.L., Lacher, D.W., Tarr, P.I., Acheson, D.W. and Whittam, T.S. 2002. Molecular characterization of a serotype O121:H19 clone, a district shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70, 6853–6859; doi:10.1128/IAI.70.12.6853-6859.2002 Tran, A., Alby, K., Kerr, A., Jones, M. and Gilligan, P.H. 2015. Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 53, 2473–2479; doi:10.1128/JCM.00833-15 | ||
| How to Cite this Article |
| Pubmed Style Biswas U, Mallick A, Das S. Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. J Microbiol Infect Dis. 2023; 13(4): 171-174. doi:10.5455/JMID.2023.v13.i4.1 Web Style Biswas U, Mallick A, Das S. Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. https://www.jmidonline.org/?mno=155274 [Access: April 27, 2024]. doi:10.5455/JMID.2023.v13.i4.1 AMA (American Medical Association) Style Biswas U, Mallick A, Das S. Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. J Microbiol Infect Dis. 2023; 13(4): 171-174. doi:10.5455/JMID.2023.v13.i4.1 Vancouver/ICMJE Style Biswas U, Mallick A, Das S. Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. J Microbiol Infect Dis. (2023), [cited April 27, 2024]; 13(4): 171-174. doi:10.5455/JMID.2023.v13.i4.1 Harvard Style Biswas, U., Mallick, . A. & Das, . S. (2023) Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. J Microbiol Infect Dis, 13 (4), 171-174. doi:10.5455/JMID.2023.v13.i4.1 Turabian Style Biswas, Urmy, Abhi Mallick, and Surojit Das. 2023. Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. Journal of Microbiology and Infectious Diseases, 13 (4), 171-174. doi:10.5455/JMID.2023.v13.i4.1 Chicago Style Biswas, Urmy, Abhi Mallick, and Surojit Das. "Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach." Journal of Microbiology and Infectious Diseases 13 (2023), 171-174. doi:10.5455/JMID.2023.v13.i4.1 MLA (The Modern Language Association) Style Biswas, Urmy, Abhi Mallick, and Surojit Das. "Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach." Journal of Microbiology and Infectious Diseases 13.4 (2023), 171-174. Print. doi:10.5455/JMID.2023.v13.i4.1 APA (American Psychological Association) Style Biswas, U., Mallick, . A. & Das, . S. (2023) Molecular detection of Escherichia coli isolated from human, animal and environmental samples targeting mdh gene: A simple one health approach. Journal of Microbiology and Infectious Diseases, 13 (4), 171-174. doi:10.5455/JMID.2023.v13.i4.1 |