| Research Article | ||
J. Microbiol. Infect. Dis., (2023), Vol. 13(2): 72–81 Original Research Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strainsYanet Pintos-Saavedra1†, Lissette Pérez-Santos1*†, Vivian Kourí-Cardellá1, Melissa Méndez González1, Jaime Pintos2, Jorge Perez3, Carlos Fonseca Gómez3, Elias Guilarte1, Yudira Soto Brito1, Yoanna Baños Morales1 and Karla Fernandez11Virology Department, Institute of Tropical Medicine “Pedro Kourí,” Havana, Cuba 2Provincial Department of STI/HIV/AIDS and Hepatitis, La Habana, Cuba 3Medical Care Department, Institute of Tropical Medicine “Pedro Kourí,” Havana, Cuba †These authors contributed equally to this work. *Corresponding Author: Lissette Pérez-Santos. Virology Department, Institute of Tropical Medicine “Pedro Kourí,” Havana, Cuba. Email: lissette [at] ipk.sld.cu Submitted: 09/01/2023 Accepted: 17/06/2023 Published: 30/06/2023 © 2023 Journal of Microbiology and Infectious Diseases
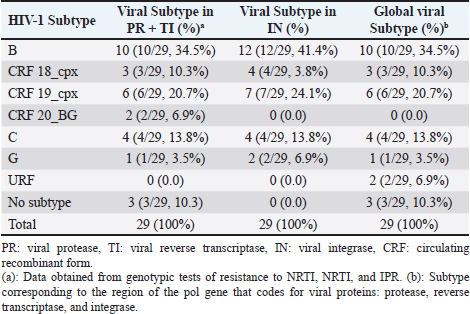
AbstractBackground: Cuban studies show high levels of viral resistance in treated and untreated patients. In 2017, dolutegravir (DTG) was introduced as a rescue therapy in Cuba, and in 2018 as a first-line regimen. The aim of this study was to determine the primary and secondary resistance of HIV-1 to DTG in Cuban patients with multiple resistances. Methods: The sample consisted of 29 patients, of which 23 had not been treated with DTG. The viral RNA isolated from plasma was amplified and the fragment of the pol gene encoding viral integrase (3,854–5,981 bp) was sequenced. The viral subtype of the 29 viruses was determined. Results: No primary resistance mutations to DTG were detected in any patient. The polymorphic positions found with greater frequency in the treated patients were: 112 and 119 (both with 73.9%), 125 (69.5%), and 201 (78.2%). Virological suppression was achieved in 18 out of 23 patients (78.2%) and in 14 out of 23 (60.8%) at 3 and 6 months after the start of treatment, respectively. Of the six patients who had received DTG prior to this study, four (66.7%) developed resistance to integrase inhibitors. The subtypes found were: B (41%), CRF19_cpx (24%), CRF18_cpx and C (both 14%), and G (7%). Conclusion: Primary resistance to DTG was not detected, while secondary resistance was high in those patients who received a single daily dose of the antiviral. It is necessary to delve into the role of some polymorphisms, more frequent in certain subtypes, and their contribution to primary resistance. Keywords: HIV, Integrase inhibitors, Dolutegravir, Multidrug-resistant virus, Cuba. IntroductionHighly effective antiretroviral therapy has changed the treatment of HIV-1 infection (Friedland, 2019). There are five classes of antiretroviral agents (ARVs): nucleoside and nucleotide analog reverse transcriptase inhibitors (NRTI), non-nucleoside reverse transcriptase inhibitors (NNRTI), protease inhibitors (PIs), entry inhibitors (antagonists of coreceptors, fusion inhibitors, and post-fixation inhibitors), and integrase inhibitors (INI) (InfoSida, 2019). As of December 2019, 33,586 people were diagnosed with HIV-1 infection in Cuba. Of them, 27,354 (81.4%) are living with HIV-1 and 94% are receiving TARVAE (MINSAP, 2019). Since the beginning of antiretroviral therapy in 1986, integrase, protease, and reverse transcriptase have been the targets for antiretroviral drug development. In 2007, the first integrase inhibitor was approved: raltegravir (RAL), followed by elvitegravir (EVG), dolutegravir (DTG), and bictegravir (BIC). These compounds are also chain transfer inhibitors (InfoSida, 2019). Integrase is encoded by the HIV-1 pol gene. It has 288 amino acids and three structural domains: the terminal amino (1–49), the catalytic nucleus, (50–211), and the terminal carboxyl (212–288) (Hazuda, 2010). Good patient adherence to treatment can suppress viral replication for decades. The therapy does not eliminate HIV-1 infection, which today is considered a chronic disease, but non-compliance with treatment schedules, drug intolerance, and interactions of ARVs with other drugs that reduce blood levels of the drug below its optimal values can cause treatment failure (Afani and Gallardo, 2011). Another cause of treatment failure is infection with resistant strains. These patients have fewer therapeutic options and are at higher risk of death and complications, particularly in developing countries, where therapeutic options are more limited (Afani et al., 2010). Primary resistance occurs in patients who have not yet received ARV treatment. It occurs due to an infection with a virus that has mutations associated with resistance, currently known as transmitted resistance. From now on, both terms will be used interchangeably. Secondary resistance appears in patients who have undergone antiretroviral treatment and occurs under the selective pressure of ARV drugs (Afani et al., 2010). At the end of 2019, 38 million people were infected with HIV-1 worldwide, of which 25.4 million were receiving some ARV treatment (Onusida, 2019). These figures not only show progress in access to treatment but also underscore the effort that is still needed to reach the 90–90–90 target established by WHO and the Joint United Nations Programme on HIV and AIDS (UNAIDS) (Onusida, 2017), whose purpose is to decrease the transmission of HIV-1 and to open a path toward the elimination of the infection. In 2017, the program proposed that 90% of HIV-1 infected people receiving ARV treatment achieve viral suppression by 2020, and that this percentage should rise to 95% by 2030 (Onusida, 2017). In Cuba by 2017, 23,500 people were living with HIV; of them 4,535 (19.3%) with treatment failure. A genotypic resistance study conducted that year, in which 201 patients with treatment failure were processed, found that 6.4% (13/201) carried multidrug-resistant viruses. In 2018, the number of infected people amounted to 25,494, and 4,130 (16.2%) of them had therapeutic failure. Multidrug-resistant viruses were identified in 0.7% (1/140) of the patients who underwent genotypic tests (HIV Glasgow, 2018). According to the data from the authors’ laboratory, at the end of 2019, 27,099 people were living with HIV, 25,607 were receiving ARV treatment, and 7,093 (27.7%) had therapeutic failure, with viral loads greater than 1,000 copies/ml. Resistance trials showed that 2.4% (2/82) of patients whose viral genotypes were studied had resistance to all antiretroviral families. For these patients, the therapeutic options are limited (Pérez et al., 2017; Kourí, 2019) and drugs that are not produced in the country are needed. In addition, HIV-pretreatment studies have found high levels of transmitted resistance in Cuba (12.5% and 29.8%) (Perez et al., 2013; Machado et al., 2019). For this reason, in 2017, Cuba decided to acquire new drugs, including DTG. Initially, this drug was used in patients with multiple resistance; as of 2018, according to WHO recommendations, it was included in the first line of treatment (Onusida, 2017). The objective of this study was to determine the transmitted (primary) resistance and secondary resistance of HIV-1 to DTG in Cuban patients with multiple resistance who used the antiviral as rescue therapy (treatment applied after the failure of at least two lines of antiretroviral treatment). Materials and MethodsSample population and inclusion criteriaPatients infected with viruses were resistant to at least one member of each of the three families of ARVs (NRTI, NNRTI, and IPR) according to the genotypic resistance test, treated at the “Pedro Kourí” Institute of Tropical Medicine in Havana, Cuba, from the beginning of January 2017 to the end of January 2019 and who agreed to participate in the study. The studied population consisted of 29 patients. At the beginning of this study, only six were under DTG treatment, and the remaining 23 patients incorporated the drug at that time. Before the initiation of the integrase inhibitor therapy, plasma samples were collected from all patients included in this study, including the six who were already under DTG treatment. Genotyping confirmed that none had primary resistance to INI. The response to therapy of the 23 patients who started treatment in the framework of this study was monitored by quantifying viral load at 3 and 6 months after initiation. If viral load values above 1,000 copies/RNA were obtained, a second test for resistance to INI was indicated. Data collectionA database was prepared considering the data obtained from the clinical history or the SIDATRAT database (Machado et al., 2019). The variables analyzed were the collection date of the plasma sample; diagnosis date; date of treatment initiation; duration of antiretroviral therapy; date of initiation of DTG treatment; drug dose used; duration of antiretroviral therapy with DTG; HIV mutations associated with resistance to PIs and reverse transcriptase; HIV mutations associated with resistance to INI; levels of resistance to INI; HIV subtypes in the sequence of the gene that encodes the proteins protease, reverse transcriptase, and integrase; viral load at the time of sample collection, and three and 6 months after starting treatment with DTG; and CD4+ cell counts at the time of sample collection. Obtaining viral RNA, processing samples, and synthesis of the cDNAOne milliliter of plasma from each patient was centrifuged at 14,000 rpm for 1 hour at 4°C. The pellet was suspended in 140 µl of the supernatant. The manual extraction of the viral RNA was carried out, according to the protocol described by the manufacturer, using the commercial QIAamp Viral RNA Mini Kit (QIAGEN, Germany). The extracted RNA was stored at −80°C until use. The synthesis of the cDNA was carried out according to the protocol described by van Laethema et al. (2008). Analysis and purification of nested PCR productsTen microliters of the sample were taken and analyzed in the QIAxcel Advanced System (QIAGEN, Germany), using the QIAxcel DNA Screening Kit (2400), the alignment pattern QX 15 bp–3 kb, and the 50 bp–1.5 kb molecular weight standard. The nested PCR products were purified with the QIAquick® PCR Purification Kit (QIAGEN, Germany), according to the protocols of the commercial house. Nucleic acid sequencingA 1,254-bp fragment of the pol gene was sequenced. This fragment spans 867 nucleotides (289 amino acids) of HIV-1 integrase. Four primers were used for each purified nested PCR product, with the aim of superimposing the sequences obtained from the different primers of the integrase region. The mixes from each sequencing reaction contained 8 µl of the dye terminator cycle sequencing (DTCS) Quick Star Master Mix DTCS Quick Start Kit (Beckman Coulter, USA), 6 µl H2O, 1 µl each of the primers, and 5 µl of purified DNA, obtaining a final reaction volume of 20 µl. Fifty cycles of 96°C for 30 seconds, 50°C for 20 seconds, and 60°C for 4 minutes. The products were purified according to the protocol described in the commercial DTCS Quick Star Master Mix kit (Beckman Coulter, EU) and were analyzed in a Beckman Coulter model CEQTM8800 automatic sequencer using the raw data analysis procedure for PCR products. The primers used were described by van Laethem et al. (2008). Sequence editing, analysis of resistance mutations to INIThe sequences obtained were assembled and edited using the Sequencher™ Version 4.10.1 computer tool (Genes Codes Corporation, USA). As a reference nucleotide sequence, the integrase gene of the HIV-1 strain B.FR.83.HXB2_LAI_IIIB_BRU.K03455 was used. Resistance-conferring mutations to integrase inhibitors and levels of resistance were identified using the Stanford University Database Genotypic Resistance Interpretation Algorithm, version 8.8 (https://hivdb.stanford.edu/hivdb / by-mutations). Determination of the HIV-1 subtypeHIV-1 subtypes were determined with the Rega subtyping tool version 3 (Pineda-Pena et al., 2013) and confirmed by manual phylogenetic analysis using CLUSTAL-X (http://www.clustal.org/clustal2/) and the binding method by neighborhood in MEGA version 5 (Kimura’s 2-parameter correction, bootstrap 1,000) (Tamura et al., 2011). The recombinant forms were assigned using the Simplot version 2.5 bioinformatics program (https://sray.med.som.jhmi.edu/SCRoftware/simplot/) (Salminen et al., 1995). CD4+ cell count and determination of viral loadThe CD4+ cell count was performed with a Becton Dickinson flow cytometer (Bio-Sciences, USA). The viral load in plasma was quantified with the NucliSENS technology (Biomérieux, France). Ethical approvalThe research was approved by the Ethics Committee of the “Pedro Kourí” Institute of Tropical Medicine. ResultsDetection of subtypes in the gene segment encoding HIV-1 integrase and analysis of the global subtypeIn the amplified samples of the 29 patients, the following viral subtypes were identified in decreasing order of frequency of appearance: subtype B, followed by CRF19_cpx, CRF18_cpx, and C subtypes in equal proportion, and lastly subtype G. The analysis of the global viral subtype (subtypes found in the protease, reverse transcriptase, and integrase proteins) showed that those found in PR-TI coincided with the subtype detected in the amplified fragment of the viral IN, except for two viruses of the recombinant form BG (CRF20_BG), one subtype G and the other, subtype B (Table 1). Table 1. Subtypes detected by partial sequencing of two regions of HIV-1 in the analyzed samples.
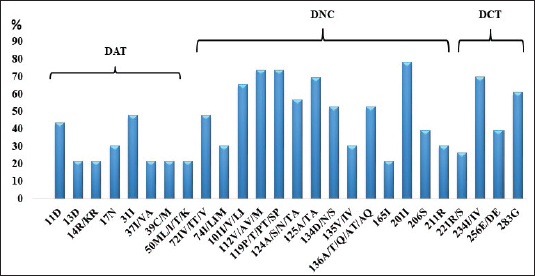
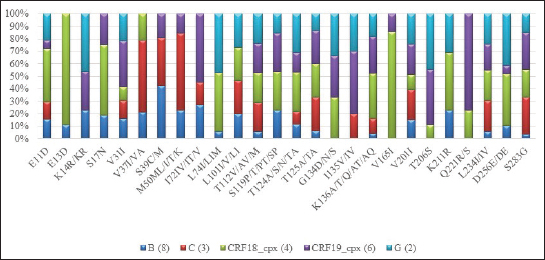
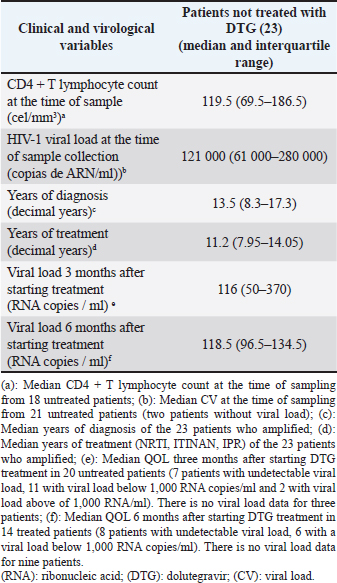
Variability of the region that encodes HIV-1 integraseIn the viral sequences of the 23 patients who had not received DTG prior to the study, polymorphisms were detected in 85 (29.5%) of the 288 amino acids of the integrase gene (Fig. 1). The most variable positions (in more than 20% of the analyzed sequences) were in the amino-terminal domain, 11 (in 43.4% of the patients) and 31 (47.8%); in the catalytic core, 112 (73.9%), 119 (73.9%), 125 (69.5%), and 201 (78.2%); at the carboxyl-terminal, 234 (69.6%) and 283 (in 60.9% of patients). Neither of these positions is associated with resistance to integrase inhibitors (Databases, 2019). In all viral subtypes, changes in amino acids were detected at positions 11, 31, 112, 124, 125, 136, 201, 234, and 283 (Fig. 2). The highest frequency of variation was found in positions 13 (E13D) and 165 (V165I) for the recombinant form CRF18_cpx (Fig. 2). Patients who had not been treated with DTG at the time of sample collectionVL values were undetectable in 7 patients (35%), and 11 below 1,000 RNA copies/ml (55%), at three months after starting treatment. However, values above 40,000 RNA copies/ml persisted in 2 patients (10%). There were three patients without quantification of VL at this time. The median VL 6 months after starting treatment was 118.5 (96.5–134.5) (6/23; 26.1%). VL values remained undetectable in eight (8/23; 34.8%) patients, while in nine patients (9/23; 39.1%), it was not available at this time and two of them died during the studied period (Table 2)—one due to chronic kidney failure and generalized bacterial sepsis and the second due to liver cirrhosis due to hepatitis C.
Fig. 1. Most frequent polymorphic positions in the coding region for HIV-1 IN detected in the 23 untreated patients. (DAT): Amino-terminal domain; (DNC): Domain of the catalytic core; (DCT): Carboxyl Terminal Domain.
Fig. 2. Most frequent polymorphic positions by subtypes in the coding region for HIV-1 IN detected in the 23 untreated patients. Table 2. Clinical and virological characteristics of patients not treated with DTG.
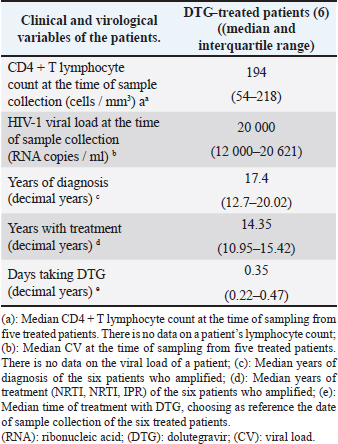
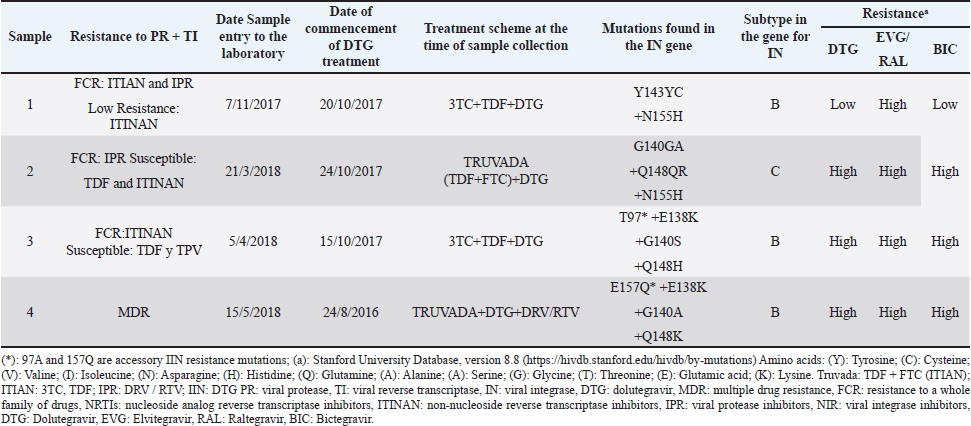
Patients who had been treated with DTG at the beginning of this studyThe genotypic test of resistance to integrase inhibitors carried out on the plasma samples collected before initiating DTG treatment from the six patients already receiving the antiretroviral showed that the amplified viruses were sensitive to the family of integrase inhibitors. In this group of patients, the time under treatment with DTG was very short (from 18 days to 6 months, with the exception of one patient who received it for 2 years) (Table 2), so the variable time of treatment with the ARV was not used in testing. These patients started the study with low CD4+ values and high VL values. The median years of HIV-1 infection and treatment with NRTI, NNRTI, or PI were 17.4 and 14.35 years, respectively (Table 3). Four patients (4/6; 66.7%) carried viruses with mutations that confer resistance to integrase inhibitors (Table 4); three of them with high resistance to the four integrase inhibitors; and one with high resistance to ELV and RAL, and low to DTG and BIC (Table 4). The patient, who was only treated with DTG for 18 days when the sample was received in our laboratory, had previously been treated with RAL. The remaining three treated patients received the drug at a dose of 50 mg/day. Table 3. Clinical characteristics of patients treated with DTG at treated with DTG at baseline
DiscussionReducing the spread of resistance is essential to obtain maximum long-term efficacy from first-line TARVAE regimens. In Kourí et al. (2012) raised the existence in Cuba of high levels of resistance (69.5%, 54.8%, and 44.4% to NRTI, NNRTI, or PI, respectively), due to prolonged exposure to failed therapies (there were limited options for antiviral substitutions), and the use of suboptimal treatment regimens using non-ritonavir-boosted PIs (Kouri et al., 2012). Since 2011, an increase in transmitted resistance has been observed, which in 2019 was manifested in 21.5% of patients, leading to a change in first-line therapies (Machado et al., 2019). Since the beginning of 2018, the new regimens used in Cuba include integrase inhibitors, a new pharmacological group with high efficacy as demonstrated in published clinical trials (Lantero et al., 2019). Table 4. Therapy history of DTG-treated patients with resistance mutations to this drug.
Detection of subtypes in the gene segment encoding HIV-1 integrase and analysis of the global subtypeFour groups of the human immunodeficiency virus are known: M, N, O, and P; M is responsible for the current pandemic. Subtypes, Circulating, and Unique Recombinant Forms of group M have been described (Kandathil et al., 2005). The wide variety of subtypes that characterizes the Cuban epidemic is due to the fact that the first-detected patients were infected on the African continent where the greatest viral variability is observed (Kourí et al., 2017; Machado et al., 2017). The viral subtype was determined in the gene segment encoding the integrase and the global subtype including the three proteins encoded by the viral polymerase gene (protease, reverse transcriptase, and integrase). In two samples, the viral subtype found in the region of the pol gene that encodes protease and the reverse transcriptase was different from that found in the region of this gene that encodes integrase. The sequence of the gene fragment encoding PR and TI corresponded to CRF20_BG, while the region of this gene encoding integrase corresponded to subtype G in one sample and subtype B in the other. The Cuban BG recombinants (CRF20_BG, CRF23_BG, and CRF24_BG) correspond to the G subtype in the fragment of the pol gene that encodes integrase, as described by Sierra et al. (2007). The other virus with subtype B in the integrase fragment could be a single recombinant, a very common phenomenon observed in geographic areas where different subtypes circulate, as in Cuba (Nájera et al., 2002). Patients not treated with DTG at the time of sample collectionThe low values of CD4+ and the high values of VL in the 23 patients not treated with DTG are related to long-term HIV-1 infection (median years of diagnosis 13.8); only one patient had been recently diagnosed. These patients received salvage therapy since they are resistant to various ARVs. Increased viral replication and decreased CD4+ levels are observed in patients with failed therapy (Korenromp et al., 2009). Three months after treatment with DTG, VL values above 40,000 RNA copies/ml persisted only in 2 of the 23 treated patients. These two patients began treatment with DTG with VL values above 100,000 copies; the high initial viral load could have made it difficult to lower this marker in such a short period of time. The median viral load at 3 and 6 months after starting DTG corroborated what has been reported in the literature on the efficacy of DTG as salvage therapy (Moreno and Berenguer, 2015). Viral load values were undetectable in 11 and 9 patients at three and 6 months, respectively. This ARV has novel characteristics, namely, a favorable pharmacokinetic profile and a prolonged intracellular half-life, which makes a single daily dose possible. It also has a high genetic barrier compared to the integrase inhibitors RAL and ELV (Dow and Bartlett, 2014). Polymorphisms in patients not treated with DTG at the time of samplingAll the polymorphic positions found in patients not treated with DTG at the beginning of this study were consistent with those published in the Stanford database (Databases, 2019). Rogers et al. (2018) observed the V31I change in South African patients infected with HIV-1 subtype C (Databases, 2019). This change occurs in 75%–85% of non-B virus sequences (Rogers et al., 2018), which is consistent with our results. The change from T to I/V/LI at position 101 was detected with frequencies above 75% in all viral subtypes, except for the CRF19_cpx subtype; while the L112V change in the wild type B subtype strain (HXB2) was observed in all non-B subtypes. Numerous changes were found in position 124 of the integrase (T by A/N/S/TA) (Fig. 2). In patients not treated with integrase inhibitors, Saladini in 2012 reported the prevalence of switching from T to A at position 124 (Saladini et al., 2012). The T125A change in integrase was found with a variable frequency in the subtypes: B (25%), G (50%), CRF19_cpx, C, CRF18_cpx (100%), which coincides with that commented by Rogers et al. (2008) who recognize this mutation as highly polymorphic in both B subtypes and non-B genetic forms. Finally, the V201I change (discussed by Rogers et al., 2018) was found with high frequency in all HIV-1 subtypes of the samples. Rogers et al. (2018) also suggest that this mutation is highly polymorphic in both B and non-B subtypes. For the success of therapies that incorporate this new family of ARVs, the possible implications of some of the polymorphisms detected should be studied because they were found in higher percentages in circulating recombinant forms that have only been described in the Cuban epidemic. Viral mutations detected in patients treated with DTG at the time of sample collectionThe mutations of resistance to integrase inhibitors detected coincide with those described (Hazuda, 2010; Wallis et al., 2017) and those published in the Stanford database (Databases, 2019). The number of patients treated with DTG at the time of sampling was very small (six patients). One of the patients treated with RAL, presented the characteristic pattern of mutations of this ARV, while others presented the characteristic pattern of those receiving treatment with DTG (Hazuda, 2010). Although in this patient no primary mutations to integrase inhibitors were found in a sample prior to treatment with DTG, it should be noted that the sequencing technique used only detects that the viral population present above 20% (Sanger method) and that in these circumstances the mutations may not be detected if pharmacological pressure is not applied. Resistance to RAL is related to the appearance of the N155H + E92Q, Q148H/R/K + G140S/A, or Y143C/R mutation patterns. EVG shares two mutation patterns with RAL: N155H + E92Q and Q148H / R / K + G140S / A (Garrido et al., 2008). Unlike RAL and ELV, multiple mutations are required to observe resistance to DTG with clinical implications: 148H/K/R/N + 138K/A/T + 140S/A/C (Wu et al., 2014). Cross-resistance between RAL and EVG is observed on the basis of mutations at positions 155 and 148. Successful treatment with DTG is not usually compromised by mutations at position 155, but by mutations at position 148 (Hazuda, 2010). High resistance to DTG requires the appearance of multiple resistance mutations to first-generation integrase inhibitors; to prevent the accumulation of mutations and preserve the response to DTG, when the therapeutic regimen is not effective, its prompt modification is of paramount importance (Anstett et al., 2017). The patient infected with a subtype C virus has high resistance to the entire family of integrase inhibitors and has the combination of the G140A, Q148R, and N155H mutations, which is consistent with the results of the VIKING study (Moreno and Berenguer, 2015). The amplified viruses in two other patients had high resistance to all integrase inhibitors. In one of them, the polymorphic accessory mutation T97A was also found, which can occur in between 1% and 5% of viral isolates from untreated patients and in patients treated with RAL and EVG (3%) (Abram et al., 2017). By itself, this mutation has minimal effects on susceptibility to integrase inhibitors, but in combination with other resistance mutations, it synergistically reduces susceptibility to EVG, RAL, DTG, and possibly to BIC (Anstett et al., 2017). The E138K mutation is found in patients receiving RAL, EVG, and DTG and usually occurs in combination with the Q148/K/R/N mutation. Its presence alone does not reduce susceptibility to integrase inhibitors; however, when it occurs in combination with the Q148 H/K/R/N mutation, it causes high resistance to RAL and EVG, and intermediate to DTG and BIC (Anstett et al., 2017). In the group of patients already treated with DTG at the time of sample collection, three viral isolates developed resistance to BIC, a drug from this family that has recently been placed on the market. An in vitro study demonstrated decreased susceptibility after the appearance of M50I/R263K mutations (Anstett et al., 2017). Likewise, in vitro studies found reduced susceptibility to BIC and cross-resistance among integrase inhibitors in viruses with a single resistance substitution at positions E92Q, T97A, Y143C/R, Q148R, and N155H (Anstett et al., 2017). G140A/C/S and Q148H/R/K substitutions, usually associated with additional mutations such as L74M, T97A, or E138A/K, showed reduced susceptibility to BIC (Anstett et al., 2017). Most authors recommend the use of DTG at a dose of 50 mg daily in patients who have not been exposed to integrase inhibitors. In these cases, a boosting agent is not required to maintain long-lasting, effective daily concentrations (Castagna et al., 2014). There are two other circumstances in which DTG has been used. First, in patients who have been previously treated with the first integrase inhibitors on the market (RAL or EVG), and who have then switched to a second scheme in which DTG is included. In this case, after previously failing RAL or EVG, the viruses had acquired new resistance mutations related to integrase inhibitors. Although the DTG dose was doubled in these cases, therapeutic failure occurred due to the preexistence of mutations that confer cross-resistance to drugs of this family (DTG, RAL, and EVG) (Moreno and Berenguer, 2015). One of the patients in our study had previously been treated with RAL; therefore, his genotypic assay showed mutations related to this ARV, which also confers low resistance to DTG. Second, DTG has been used in people infected with multi-resistant viruses, that is, they do not respond to treatments with other families of ARVs (NRTI, NNRTI, or PI). In this case, ARV doses of 50 mg twice daily have been recommended. The literature suggests that, in these cases, the probability of therapeutic success with DTG is greater than in patients previously treated with other drugs of this family, probably because these viruses do not have mutations related to integrase inhibitors (Castagna et al., 2014). This is the case of the three patients in our study in whom highly resistant viruses to DTG were detected. They started a rescue therapy with a single dose of DTG (50 mg once a day), which must have led to the rapid accumulation of mutations and the subsequent treatment failure. ConclusionPrimary resistance to DTG was not detected, while secondary resistance was high in those patients who received a single daily dose of the antiviral. It is necessary to delve into the role of some polymorphisms, which were more frequent in certain subtypes, and their contribution to primary resistance. The viral subtype in the studied fragment coincided in a high percentage with that detected in the fragment of the reverse transcriptase and the viral protease. AcknowledgmentsThe authors of the manuscript thank all the patients who agreed to participate in the study, the Heads of the Cuban STI, HIV/AIDS Program, and the United Nations Development Program (UNDP). Author contributionsYanet Pintos and Lissette Pérez: Conceptualization and design of the study, data curation, methodology, software, and writing (review and editing). Vivian Kourí: Writing (review and editing) and supervision. Melisa Mendez: Writing (review and editing). Jaime Pintos, Jorge Pérez, and Carlos Fonseca: Investigation and supervision. Elias Guilarte and Yudira Soto: Formal analysis and methodology. Yohana Baños, Karla Fernandez: Methodology. ReferencesAfani, A., Beltran, C., Maria Gallardo, A., Roessler, P., Acevedo, W. and Vasquez, P. 2010. Prevalence of primary antiretroviral resistance among HIV infected patients in Chile. Rev. Méd. Chile. 138(6), 669–676; http://doi:10.4067/s0034-98872010000600002. Afani, S. and Gallardo, O. 2011. Resistencia a la terapia antiretroviral en la infeccion por virus de inmunodeficiencia humana. Rev. Chilena. Infectol. 28(5), 461–469; http://dx.doi.org/10.4067/S0716-10182011000600011 Aragones Lopez, C., Campos Diaz, J.R., Perez Correa, D.F., Martinez Rodriguez, A., Perez Avila, L.J. 2012. Sidatrat: informatics to improve HIV/AIDS care. Med. Rev. 14(4), 5–9; http://doi: 10.37757/MR2012V14.N4.3 Anstett, K., Brenner, B. and Mesplede, T. 2017. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 14, 1742–4690; http://doi: 10.1186/s12977-017-0360-7 Abram, M.E., Ram, R.R., Margot, N.A., Barnes, T.L., White, K.L. Callebaut, C. 2017. Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PloS. One. 12(2); (http://doi: 10.1371/journal.pone.0172206) Castagna, A., Maggiolo, F., Penco, G., Wright, D., Mills, A. and Grossberg R. 2014. Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J. Infect. Dis. 210(3), 354–362; http://doi: 10.1093/infdis/jiu051 Databases. 2019. Major Integrase Inhibitor (INSTI) Resistance Mutations. Available via https://hivdb.stanford.edu/hivdb/by-mutations (Accessed 7 December 2019). Databases. 2019. Mutation prevalence according to subtype and treatment. . Available via https://hivdb.stanford.edu/hivdb/by-mutations (Accessed 7 December 2019). Dow, D.E. and Bartlett, J.A. 2014. Dolutegravir, the second-generation of integrase strand transfer inhibitors (INSTIs) for the treatment of HIV. Infect. Dis. Ther. 83–102; http://doi:10.1007/s40121-014-0029-7. Friedland, G.H. 2019. Early treatment for HIV: the time has come. N. Engl. J. Med. 322, (14) 1000–1002; https://doi:10.1056/NEJM199004053221409. Garrido, C., de Mendoza, C. and Soriano, V. 2008. Resistance to integrase inhibitors. Enfermedades. Infecc. Y. Microbiol. Clin. 26 Suppl 12, 40–46. Hazuda DJ. 2010. Resistencia a los inhibidores de la integración del Virus de la inmunodeficiencia humana tipo I. Actual. Sida. 18(70), 135–141; https://doi.org/10.1016/S1413-8670(10)70103-3 HIV Glasgow. 2018. 2018, 28–31 October 2018, Glasgow, UK. J. Int. AIDS Soc. 21(Suppl 8),1758–2652. InfoSida. 2019. Available via. https://infosida.nih.gov/understanding-hiv-aids (Accessed 7 September 2019). Kandathil, A.J., Ramalingam, S., Kannangai, R., David, S. and Sridharan, G. 2005. Molecular epidemiology of HIV. Indian. J. Med. Res. 2121(4), 333–344. Kourí, V. Actualización de las ITS y en Resistencia Antirretroviral Retos en el Tratamiento de pacientes con VIH/sida: Experiencia cubana; La Habana, Cuba 2019. Kouri, V., Aleman, Y., Pérez, L., Pérez, J., Fonseca, C. and Correa, C. 2012. High frequency of antiviral drug resistance and non-B subtypes in HIV-1 patients failing antiviral therapy in Cuba. J. Clin. Virol. 55(4), 348–355; http://doi: 10.1016/j.jcv.2012.08.019 Kourí, V., Alemán, Y., Pérez, L., Martínez, Y., Pérez, J. and Fonseca, M. 2017. Surveillance of HIV-1 subtype and antiretroviral resistance in Cuban individuals failing therapy during 2009-2016. J. Int. AIDS. Soc. 20(Supl 2), 41–42; http:// Korenromp, E.L., Williams, B.G., Schmid, G.P. and Dye, C. 2009. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection--a quantitative review. PloS. One. 4(6), e5950; http://doi:10.1371/journal.pone.0005950. Lantero Abreu, M.I., Sánchez Fuentes, J., Fiol, J.J., Betancourt Llody, Y.A., Cancio Enrique, I. and Matos Morejón, M.J. 2019. Plan estrategico nacional para la prevención y control de las ITS, el VIH y las hepatitis 2019 - 2023. 2019. Machado, L.Y., Pintos, Y., Díaz, H.M., Pérez, L., Blanco, M. and Kourí, V. 2017. Increase of recombinant forms CRF20, 23, 24_BG and Several URF of HIV-1 among newly diagnosed Cuban patients: 2013-2014. ARC. J. AIDS. 2(1), 24–31. Machado, L.Y., Blanco, M., López, L.S., Díaz, H.M., Dubed, M. and Valdés, N. 2019. National survey of pre-treatment HIV drug resistance in Cuban patients. PloS. one. 14(9), e0221879; http://doi:10.1371/journal.pone.0221879 MINSAP. 2019. Base Nacional de Datos de la Infección por VIH. Anual. Cuba2019. Moreno, S. and Berenguer, J. 2015. Efficacy of dolutegravir in treatment-experienced patients: the SAILING and VIKING trials. Enferm Infecc Y Microbiol Clin. 33 Suppl 1, 26–30; http://doi:10.1016/S0213-005X(15)30006-9 . Nájera, R., Delgado, E., Pérez-Alvarez, L. and Thomson, M.M. 2002. Genetic recombination and its role in the development of the HIV-1 pandemic. AIDS. 16 Suppl 4:S3–16; http://doi:10.1097/00002030-200216004-00002. Laethema, K.V., Schrootena, Y., Covensa, K., Dekeersmaekera, N., Munterc, P.D. and Wijngaerdenc, E.V. 2008. A genotypic assay for the amplification and sequencing of integrase from diverse HIV-1 group M subtypes. J. Virol. Methods. 153, 176–181; http://doi:10.1016/j.jviromet.2008.07.008 Onusida. 2019. El Sida en cifras. Available via http://aidsinfo.unaids.org (Accessed 7 December 2019). Onusida. 2017. Abordaje de la farmacorresistencia del VIH: Tendencias, Directrices y Acción Mundial. 2017. [http://aidsinfo.unaids.org, cited 2019]. Pérez, L., Kourí, V., Alemán, Y., Correa, C., Martínez, Y. and Pérez, J. 2017. Mecanismos de Resistencia del VIH a las drogas antirretrovirales. Vigilancia de la resistencia antirretroviral del VIH-1 en Cuba en paciente con fallo a la terapia durante el periodo 2009-2017. Congreso 80 aniversario del IPK; La Habana, Cuba 2017. Perez, L., Kouri, V., Aleman, Y., Abrahantes, Y., Correa, C. and Aragones, C. 2013. Antiretroviral drug resistance in HIV-1 therapy-naive patients in Cuba. Infect. Genet. Evol. 16, 144–50; http://doi:10.1016/j.meegid.2013.02.002. Pineda-Pena, A.C., Faria, N.R., Imbrechts, S., Libin, P., Abecasis, A.B. and Deforche, K. 2013. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect. Genet. Evol. 19, 337–348; http://doi:10.1016/j.meegid.2013.04.032 Rogers, L., Obasa, A.E., Jacobs, G.B., Sarafianos, S.G., Sönnerborg, A. and Neogi, U. 2018. Structutal implications of genotypic variations in HIV-1 integrase from diverse subtypes. Front Microbiol. 9, 1754; http://doi:10.3389/fmicb.2018.01754 . Salminen, M.O., Carr, J.K., Burke, D.S. and McCutchan, F.E. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS. Res. Hum. Retroviruses. 11(11), 1423–1425; http:/doi: 10.1089/aid.1995.11.1423/ Sierra, M., Thomson, M.M., Posada, D., Perez, L., Aragones, C. and Gonzalez, Z. 2007. Identification of 3 phylogenetically related HIV-1 BG intersubtype circulating recombinant forms in Cuba. J. Acquir. Immune. Defic. Synd. (1999). 45(2), 151–160; http://doi:10.1097/QAI.0b013e318046ea47 Saladini, F., Meini, G., Bianco, C., Monno, L., Punzi, G. and Pecorari, M. 2012. Prevalence of HIV-1 integrase mutations related to resistance to dolutegravir in raltegravir naive and pretreated patients. Clin. Microbiol. Infect. Dis. 18(10), E428–E430; http://doi:10.1111/j.1469-0691.2012.03917.x. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28(10), 2731–2739; http://doi:10.1093/molbev/msr121 Wallis, C.L., Viana, R.V., Saravanan, S., Silva de Jesus, C., Zeh, C. and Halvas, E.K. 2017. Performance of Celera RUO integrase resistance assay across multiple HIV-1 subtypes. J. Virol. Methods. 241, 41–45; http://doi:10.1016/j.jviromet.2016.12.008 Wu, G., Abraham, T. and Saad, N. Dolutegravir for the treatment of adult patients with HIV-1 infection. Exp. Rev. Anti-Infect. Ther. 12(5),535–544; http://doi:10.1586/14787210.2014.907525 | ||
| How to Cite this Article |
| Pubmed Style Pintos-saavedra Y, Pérez L, Kourí-cardellá V, González MM, Pintos J, Ávila JP, Gomez CF, Guilarte E, Brito YS, Morales YB, Fernandez K. Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. J Microbiol Infect Dis. 2023; 13(2): 72-81. doi:10.5455/JMID.2023.v13.i2.4 Web Style Pintos-saavedra Y, Pérez L, Kourí-cardellá V, González MM, Pintos J, Ávila JP, Gomez CF, Guilarte E, Brito YS, Morales YB, Fernandez K. Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. https://www.jmidonline.org/?mno=302657414 [Access: July 15, 2025]. doi:10.5455/JMID.2023.v13.i2.4 AMA (American Medical Association) Style Pintos-saavedra Y, Pérez L, Kourí-cardellá V, González MM, Pintos J, Ávila JP, Gomez CF, Guilarte E, Brito YS, Morales YB, Fernandez K. Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. J Microbiol Infect Dis. 2023; 13(2): 72-81. doi:10.5455/JMID.2023.v13.i2.4 Vancouver/ICMJE Style Pintos-saavedra Y, Pérez L, Kourí-cardellá V, González MM, Pintos J, Ávila JP, Gomez CF, Guilarte E, Brito YS, Morales YB, Fernandez K. Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. J Microbiol Infect Dis. (2023), [cited July 15, 2025]; 13(2): 72-81. doi:10.5455/JMID.2023.v13.i2.4 Harvard Style Pintos-saavedra, Y., Pérez, . L., Kourí-cardellá, . V., González, . M. M., Pintos, . J., Ávila, . J. P., Gomez, . C. F., Guilarte, . E., Brito, . Y. S., Morales, . Y. B. & Fernandez, . K. (2023) Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. J Microbiol Infect Dis, 13 (2), 72-81. doi:10.5455/JMID.2023.v13.i2.4 Turabian Style Pintos-saavedra, Yanet, Lissette Pérez, Vivian Kourí-cardellá, Melissa Méndez González, Jaime Pintos, Jorge Pérez Ávila, Carlos Fonseca Gomez, Elias Guilarte, Yudira Soto Brito, Yoanna Baños Morales, and Karla Fernandez. 2023. Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. Journal of Microbiology and Infectious Diseases, 13 (2), 72-81. doi:10.5455/JMID.2023.v13.i2.4 Chicago Style Pintos-saavedra, Yanet, Lissette Pérez, Vivian Kourí-cardellá, Melissa Méndez González, Jaime Pintos, Jorge Pérez Ávila, Carlos Fonseca Gomez, Elias Guilarte, Yudira Soto Brito, Yoanna Baños Morales, and Karla Fernandez. "Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains." Journal of Microbiology and Infectious Diseases 13 (2023), 72-81. doi:10.5455/JMID.2023.v13.i2.4 MLA (The Modern Language Association) Style Pintos-saavedra, Yanet, Lissette Pérez, Vivian Kourí-cardellá, Melissa Méndez González, Jaime Pintos, Jorge Pérez Ávila, Carlos Fonseca Gomez, Elias Guilarte, Yudira Soto Brito, Yoanna Baños Morales, and Karla Fernandez. "Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains." Journal of Microbiology and Infectious Diseases 13.2 (2023), 72-81. Print. doi:10.5455/JMID.2023.v13.i2.4 APA (American Psychological Association) Style Pintos-saavedra, Y., Pérez, . L., Kourí-cardellá, . V., González, . M. M., Pintos, . J., Ávila, . J. P., Gomez, . C. F., Guilarte, . E., Brito, . Y. S., Morales, . Y. B. & Fernandez, . K. (2023) Primary and secondary resistance of HIV-1 to integrase inhibitors in Cuban patients infected with multidrug-resistant HIV strains. Journal of Microbiology and Infectious Diseases, 13 (2), 72-81. doi:10.5455/JMID.2023.v13.i2.4 |