| Research Article | ||
J. Microbiol. Infect. Dis., (2024), Vol. 14(2): 46–54 Research Article Growth pattern, protein quantification, and antibiotic responses of four halophilic bacterial strains in different salt concentrationAnkur Das1* and Mahua Gupta Choudhury21Department of Microbiology, Assam Don Bosco University, Guwahati, India 2Department of Biotechnology, Assam Don Bosco University, Guwahati, India *Corresponding Author: Ankur Das. Department of Microbiology, Assam Don Bosco University, Guwahati, India. Email: nkrdas2 [at] gmail.com Submitted: 25/02/2024 Accepted: 04/06/2024 Published: 30/06/2024 © 2024 Journal of Microbiology and Infectious Diseases
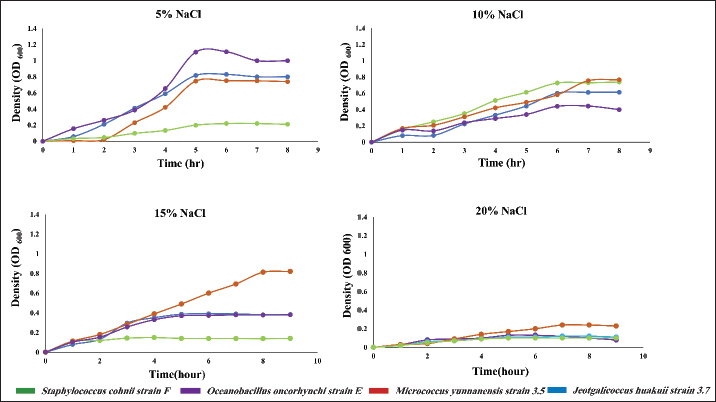
ABSTRACTBackground: Halophilic bacteria are studied extensively to explore molecular basis of adaptation, enzymatic potential, drug resistance and their other biotechnological implications. Their ability to survive in hypersaline condition is often linked to the potential of biosynthesizing a well-known compatible solute, namely ectoine. Aim: In this study, four bacterial strains, which were previously identified from hypersaline environment were studied for their growth pattern and ectoine biosynthesis protein expression under varying NaCl concentrations. In addition, antibiotic susceptibility of the strains was also investigated. Methods: The growth pattern of the bacterial strains were studied in Luria-Bertani broth supplemented with different NaCl concentrations. Bradford method was employed for protein estimation, and optical densities of the samples were quantified through UV-Vis spectrophotometer. Moreover, expression of the proteins responsible for biosynthesis of ectoine were detected through SDS-PAGE, and disc diffusion method was used for profiling the antibiotic resistance of the strains. Results: The results indicated distinct growth patterns, with each strain demonstrating preferences for specific salinity levels. Notably, Staphylococcus cohnii strain F exhibited optimal growth at 10% NaCl, while Jeotgalicoccus huakuii strain 3.7 thrived at 15% NaCl. Total protein analysis revealed a consistent trend of decreasing protein content with rising NaCl concentrations, elucidating a potential adaptive strategy. Staphylococcus cohnii strain F stood out, maintaining higher protein levels at 20% NaCl, indicative of unique adaptive mechanisms. Antibiotic susceptibility testing uncovered diverse resistance profiles, emphasizing the strain-specific nature of responses. The expression of ectoine biosynthesis genes in S. cohnii strain F underlines its adaptive advantage in high salinity. Conclusion: The findings contribute not only to basic microbial physiology knowledge but also hold implications for biotechnological applications and clinical practices, emphasizing the importance of tailored approaches in antibiotic therapy and highlighting the strain-specific nature of bacterial adaptations. Keywords: Halophilic bacteria, Salinity, Protein expression, Antibiotic susceptibility. IntroductionHalophilic microorganisms can be found extensively across the domains of bacteria, archaea, and eukaryote, showcasing an impressive ability to thrive in hypersaline environments such as salterns, salt lakes, and seawater (Oren et al., 2002; Thombre et al., 2016). These microorganisms exhibit resilience to salt concentrations ranging from 0.2 to 5 molL-1. They are notably abundant in products like fishes and hides and find extensive uses in various industrial applications (Dutta et al., 2022). To navigate the challenges of hypersaline environments, halophiles employ strategies such as biosynthesis and secretion of solutes like ectoine, betaine, pigments, and enzymes. These mechanisms play a crucial role in maintaining osmotic balance and preventing protein denaturation (Boltyanskaya et al., 2005). Ectoine, in particular, has emerged as a remarkable solute for halophilic bacteria and was first identified in the halophilic phototrophic species Halorhodospora halochloris (Galinski et al., 1993). The biosynthesis of ectoine involves several steps. The precursor molecule, aspartate semialdehyde, an intermediate in amino acid metabolism, is converted to L-2,4-diaminobutyric acid. This compound is then acetylated to form Nγ-acetyldiaminobutyric acid, which undergoes cyclization to produce ectoine (Nakayama et al., 2000; Wei et al., 2011). The genes responsible for ectoine biosynthesis, namely ectA, ectB, and ectC, have been identified and characterized. The gene ectA encodes diaminobutyric acid acetyltransferase, ectB encodes diaminobutyric acid aminotransferase, and ectC encodes ectoine synthase (Kang et al., 2022). These enzymes have been studied in detail, revealing insights into their activation by NaCl, suggesting a partial regulation of ectoine accumulation by intracellular cations. The comprehensive understanding of these molecular processes not only advances our knowledge of extremophiles but also holds promise for applications in various biotechnological and industrial contexts. Over the past few decades, the issue of drug resistance has evolved into a significant problem, extending beyond pathogenic infections to encompass non-pathogenic environmental strains, particularly in halophiles found in coastal seas and deep ocean waters (Thombre et al., 2016). The introduction of sewage, wastewater, and residues from healthcare facilities into saline environments has notably contributed to the proliferation of resistance, primarily among halophilic organisms (Fouz et al., 2020, Mehanni et al., 2023). Instances of resistant strain dissemination have been documented, with reports of Escherichia coli, Klebsiella pneumoniae, Pseudomonas spp., and Salmonella spp. exhibiting drug resistance in the saline waters of Mumbai possibly due to horizontal gene transfer (Vaidya and Manzoor 2009). Moreover, in America, Vaidya et al., (2011) highlighted river water containing strains producing extended spectrum β-lactamase. The study of drug resistance in halophilic microbes has revealed the presence of resistant genes acquired through horizontal gene transfer (Da Costa et al., 2013). An example is the plasmid pMH1 (11.5 kbp) isolated and characterized from H. elongata, conferring resistance to various antibiotics, including tetracycline, kanamycin, and neomycin (Fernandez-Castillo et al., 1992). Shinde and Thombre (2016) demonstrated that there is a continuous increase in the drug resistance in the aquatic saline strains likely due to dissemination of resistance strains from wastewater and sewage in the seas. The identification of pathogenic and antibiotic-resistant halophilic microorganisms underscores the importance of investigating environmental strains to comprehend the current spectrum of resistance in these bacterial populations. This research is crucial for informing strategies to mitigate the spread of resistance in diverse ecosystems. The current investigation delves into the analysis of halophilic bacteria, which were initially isolated from the Arabian Sea in Maharashtra, India, as documented in Das et al., (2019). Specifically, this study focuses on exploring the expression of ectoine biosynthesis genes in conditions of hyper salinity. Simultaneously, the isolated bacteria were subjected to screening against eleven different antibiotics. The research sheds light on the salt and antibiotic tolerance exhibited by these halophilic bacteria, providing valuable insights into their adaptability to hypersaline environments. Materials and MethodsRetrieval of bacterial isolates and growth pattern studyThe bacterial isolates, namely Oceanobacillus oncorhynchi strain E (MH591773), Jeotgalicoccus huakuii strain 3.7 (MH591772), Micrococcus yunnanensis strain 3.5 (MH591771), and Staphylococcus cohnii strain F (MH591774), which had been previously identified and stored at -80°C, were brought back to active cultures from glycerol stocks. To assess their growth patterns, fresh cultures of these strains were initiated in modified Luria-Bertani (LB) broth supplemented with varying concentrations of salt (Alam et al. 2021). 1 loop full of culture was grown overnight in 15 ml of broth, and 2% from this stock was used as inoculum size for subsequent experiments. The optical density (OD) of each bacterial culture was measured at different time points, and the resulting data were used to generate OD versus time graphs for each isolate. The doubling time of the isolates at different salt concentration was calculated using the formula y=Aebx following per method describe by Rohim et al. (2020). Protein isolation and estimationTotal protein extraction from bacterial cells followed a modified protocol based on Choudhury and Saha (2012). The lysis buffer, consisting of 65 mM Tris pH (6.8–7.4), 2% SDS, 10% glycerol, and 0.1% Triton X–100, was used in the process. The bacterial isolates were cultivated in media containing NaCl until reaching an OD of 0.5 at 600 nm. Subsequently, 5 ml of each culture was centrifuged at 16,000 rpm for 5 minutes, and the supernatant was discarded. The resulting pellet was resuspended in 100 µl of cold lysis buffer. A series of steps, including boiling at 95°C and freezing at −20°C, were iteratively performed over 10 minutes to facilitate cell lysis. A small fraction of the lysate was used to assess the protein concentration, while the remaining samples were stored at −20°C for further analysis. For protein estimation, the Bradford method was employed, following the procedure outlined by Bradford in 1976. A stock solution of bovine serum albumin (BSA) at a concentration of 1 mg/ml was prepared and subsequently diluted. OD readings for all samples were recorded using a spectrophotometer (Model: PC-based double beam spectrophotometer 2202). A standard graph was constructed with BSA concentration (µg/ml) on the X-axis and OD on the Y-axis. This method was repeated to determine the protein concentrations of cells grown in media containing 5%, 10%, 15%, and 20% sodium chloride. Separation and visualization of protein using SDS-PAGESDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis) was conducted to separate the isolated proteins following the methodology outlined by Choudhury and Saha (2012). The gel preparation involved a 12% resolving gel and an 8% stacking gel. A 10 µl volume of the protein mixture was taken, to which 10 µl of loading buffer was added, followed by the addition of 1 µl of bromophenol blue as a loading dye. The electrophoresis chamber was connected to the power supply, and the voltage was set to 200V for 60 minutes. For visualization, a staining solution was prepared using Coomassie blue R350 in 40% (v/v) methanol in water. After the electrophoresis run, gels were washed with distilled water, followed by staining overnight. The next day, the gels were destained until clear, visible bands were observed. This process allows for the separation and visualization of proteins based on their molecular weight. Antibiotic sensitivity assayThe antibiotic resistivity test for the four isolates was conducted using a susceptibility test disk method. Various antibiotics were employed for the study, including: Amoxyclave (AMC, 30 mcg); Linezolid (LZ, 30 mcg); Tetracycline (TE, 30 mcg); Ciprofloxacin (5 mcg); Ceftazidime (CAZ, 30 mcg); Chloramphenicol (C, 30 mcg); Imipenem (IPM, 10 mcg); Clavulanic (CAC, 10 mcg); Methicillin (MET, 5 mcg); Amikacin (AK, 30 mcg), and Gentamicin (GEN, 10 mcg). The procedure involved spreading cells on an LBA (Luria-Bertani Agar) plate supplemented with 5% sodium chloride, followed by the placement of antibiotic disks on the plate. Subsequently, the plates were incubated at 37˚C for 24 hours, and the zones of inhibition were measured. This method helps determine the susceptibility of the bacterial isolates to various antibiotics by observing the clear zones around the antibiotic disks, indicating inhibition of bacterial growth. ResultsGrowth curve analysisThe growth behavior of four moderately halophilic strains viz., Oceanobacillus oncorhynchi strain E, J. huakuii strain 3.7, Micrococcus yunnanensis strain 3.5, and S. cohnii strain F were explored in LB media supplemented with NaCl concentrations ranging from 5%to 20%. These observations revealed distinctive growth patterns. In the case of O. oncorhynchi strain E, it displayed robust growth at 5% NaCl, reaching a maximum OD of 1.2 during the exponential phase at fifth hour. However, a decline in growth was observed at the eighth hour from inoculation. Similarly, J. huakuii strain 3.7, and M. yunnanensis strain 3.5 reached OD close to 0.8 at 5th hour. Staphylococcus cohnii strain F, however, exhibited limited growth at 5% NaCl, with its OD not exceeding 0.2. Interestingly, at 10% NaCl, this strain showed relatively optimal growth compared to the other strains. M. yunnanensis strain 3.5 demonstrated optimal growth at 15% NaCl, reaching an OD of 0.8 after 8 hours of inoculation. Overall, at 20% NaCl, none of the strains achieved optimal growth. The growth curves for all four strains are visually represented in Figure 1. These findings provide valuable insights into how these halophilic strains respond to different saline environments.
Fig. 1. The growth pattern of all four strains in different time point supplemented with 5%–20 % NaCl.
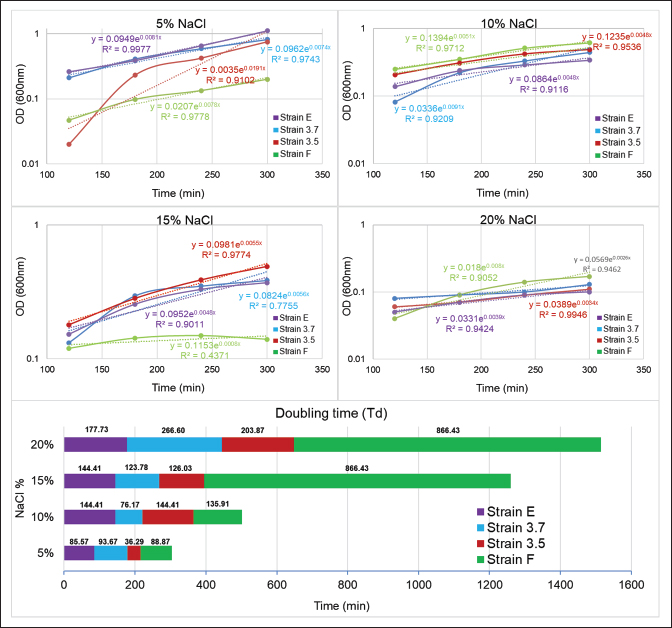
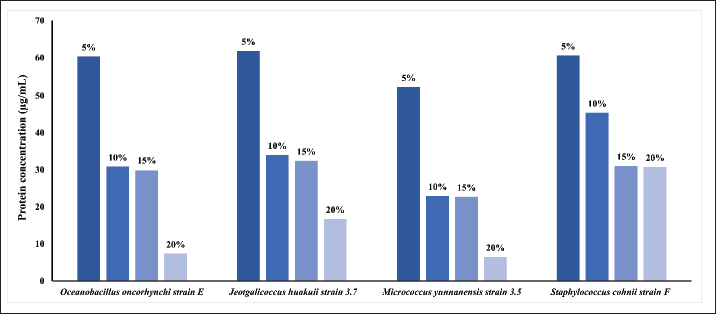
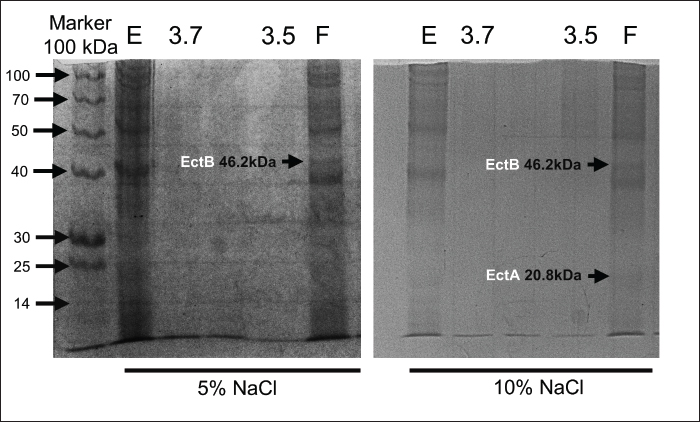
Fig. 2. Comparing doubling time of the 4 isolates in minutes at different salt concentration. Doubling time of the strains at different salt concentrationFor all the isolates the doubling time (Td) increases with increased salt concentration as presented in Figure 2. For instance, Strain E doubled at 85.57 minutes at 5% NaCl, which reached to 177.73 minutes at 20%, indicate the impact of higher salt concentration. Interestingly, strain 3.5, Td decreased at 15% to 126.03 minutes from 144.41 at 10%, indicating its high tolerance and acclimatation to higher salt amount. Protein estimation and expression of ectoine biosynthesis proteinsThe isolation and analysis of total proteins from the four isolates, grown in LB media supplemented with varying NaCl concentrations (5%, 10%, 15%, and 20%), yielded interesting insights. A comparative analysis, illustrated in Figure 3, showcased a trend where total protein content within the cells decreased with an increase in salt concentration. This observation suggests that the expression of only a limited number of proteins possibly necessary for survival in higher salt concentrations. At 5% NaCl, all isolates exhibited a total protein content of approximately 60 µg/ml. However, at 10%–15% NaCl, protein levels were estimated to be in the range of 25–30 µg/ml, with the least observed at 20% NaCl (10 µg/ml). Notably, S. cohnii strain F demonstrated a protein quantity at 20% NaCl similar to that observed at 15% NaCl, i.e., 30 µg/ml. Subsequent SDS-PAGE analysis of the total proteins revealed prominent expression of EctB (46.2 kDa) and EctA (20.8 kDa), responsible for ectoine biosynthesis, specifically in S. cohnii strain F at 10% NaCl and only EctB (46.2 kDa) at 5% NaCl (Fig. 4). Intriguingly, no expression of EctC was detected in either of the strains. Other protein bands with different molecular weights, likely associated with survival or housekeeping functions, were also observed. However, in O. oncorhynchi strain E, J. huakuii strain 3.7, and M. yunnanensis strain 3.5, no expression of ectoine biosynthesis genes was noted. The bands corresponding to other ectoine biosynthesis genes were not identified when these strains were grown in 15%–20% NaCl-supplemented media (data not showed). These findings indicate the strain-specific nature of ectoine biosynthesis under varying saline conditions.
Fig. 3. A comparative analysis showcasing a trend of total protein content in ug/ml within the cells at different salt concentration.
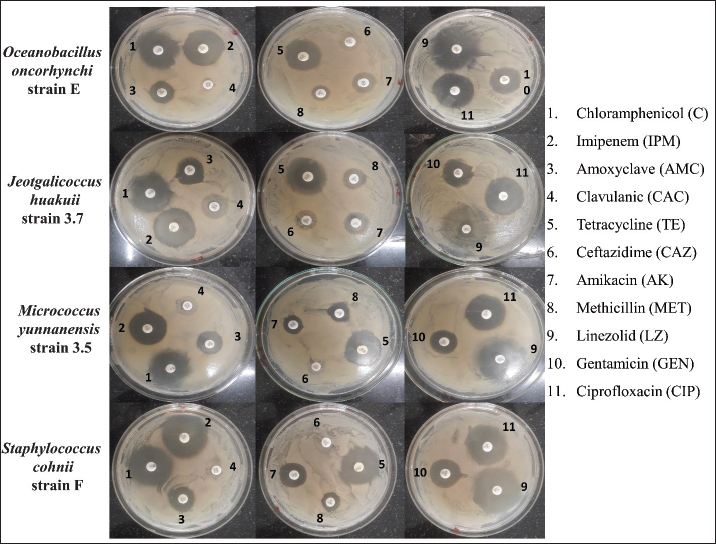
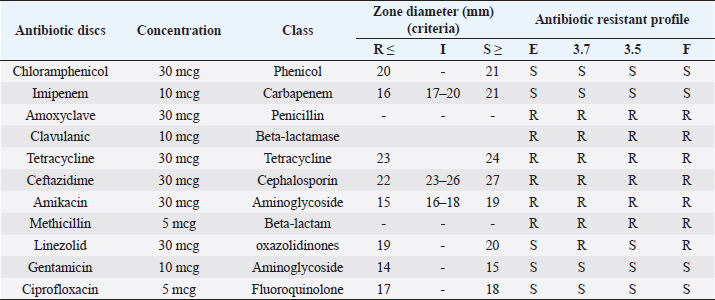
Fig. 4. SDS-PAGE analysis of the total proteins revealed prominent expression of EctB (46.2 kDa) and EctA (20.8 kDa), responsible for ectoine biosynthesis, specifically in S. cohnii strain F only at 10% NaCl and only EctB (46.2 kDa) at 5% NaCl. Antibiotic sensitivity testThe results of antibiotic susceptibility testing using the disk diffusion method for the four bacterial strains was depicted in Figure 5, and data interpretation was provided in Table 1. Each row in the table corresponds to a specific antibiotic, providing details such as the concentration of the antibiotic disc, the antibiotic class, the measured zone diameter in millimeters, and the resulting interpretation of the susceptibility profile. For example, Chloramphenicol, categorized under the Phenicol class, was tested at a concentration of 30 mcg. According to the interpretation criteria, it was classified as resistant (R), suggesting potential ineffectiveness against the tested bacterial strain, sensitive (S) indicates effectiveness of the tested antibiotic. The result suggested that most of the antibiotics including Amoxyclave, Clavulanic, Tetracycline, Ceftazidime, Amikacin, Methicillin were ineffective against all the four strains. In contract, the isolates were sensitive towards Chloramphenicol, Imipenem, Gentamicin, and Ciprofloxacin. The antibiotic resistance profile summarizes the overall susceptibility of the four bacterial strains to each antibiotic, simplifying information for clinical decision-making. The results indicated the importance of antibiotic selection based on the specific characteristics of the bacteria under investigation.
Fig. 5. The illustration of eleven antibiotic resistant profile of the four bacterial strains in LB agar plate carried out through disk diffusion method. Table 1. Antibiotic resistant profile of Oceanobacillus oncorhynchi strain E, J. huakuii strain 3.7, Micrococcus yunnanensis strain 3.5, S. cohnii strain F against 11 antibiotics of different class as per criteria guidelines of CLSI M100-S24 for disk diffusion.
DiscussionIn this study, the comprehensive analysis of the growth pattern, protein expression, and antibiotic susceptibility of the four bacterial strains viz., O. oncorhynchi strain E, J. huakuii strain 3.7, M. yunnanensis strain 3.5, and S. cohnii strain F, provides an integrated understanding of their adaptive survival under hypersaline conditions. The growth of the strains in LB media supplemented with different NaCl concentrations revealed distinctive patterns. For e.g., O. oncorhynchi strain E exhibited robust growth at 5% NaCl, emphasizing its preference for moderate salinity. Staphylococcus cohnii strain F, on the other hand, showed optimal growth at 10% NaCl, indicating its adaptation to higher salinity conditions. M. yunnanensis strain 3.5 demonstrated peak growth at 15% NaCl, showcasing its preference for even higher salt concentrations. Notably, none of the strains achieved optimal growth at 20% NaCl, suggesting a limit to their halotolerance. A total of 28 halophilic strains were isolated from Indian salterns which were able to grow in hypersaline condition producing hydrolytic enzymes for their survival (Rathakrishnan et al., 2022). Similarly, halophilic bacteria were also found to survive through metal tolerant ability in hypersaline environment (Diba et al., 2021). These findings contribute valuable insights into how these halophilic strains respond to varying saline environments, laying the groundwork for further exploration of their adaptive mechanisms and potential applications in biotechnological and environmental The analysis of total proteins further complemented the growth behavior findings. A consistent trend was observed across all strains as the NaCl concentration increased, the total protein content within the cells decreased. Staphylococcus cohnii strain F maintained relatively higher protein levels at 20% NaCl, indicating a unique adaptive strategy. SDS-PAGE analysis of total proteins revealed the expression of EctB and EctA, involved in ectoine biosynthesis, specifically in Staphylococcus cohnii strain F. Interestingly, other strains did not express these genes, emphasizing the strain-specific nature of ectoine biosynthesis. Report suggests that ectoine synthesis increased with increase in the salt concentration, and at 6% NaCl, higher concentration of ectoine was obtained (Kang et al., 2021). The use of Halomonas elongata as the primary bacterium for commercial ectoine production is well-established. However, the challenges associated with the high-salt fermentation broth used for efficient ectoine biosynthesis have prompted efforts to explore alternative chassis cells that can achieve high yields of ectoine with reduced salt levels (Fatollahi et al., 2020). The findings of this study contribute valuable information to our understanding of the molecular mechanisms underlying the adaptation of halophilic strains to saline environments, shedding light on potential targets for further exploration in the context of microbial physiology and biotechnological applications. The antibiotic susceptibility testing results underscore the diversity in resistance profiles among the strains. Different antibiotic classes exhibited distinct resistance patterns, reinforcing the need for a tailored approach in antibiotic selection. For instance, S. cohnii strain F and 3.7 showed resistance to linezolid, while E and 3.5 were sensitive to the same. However, the strains were resistance to most of the tested antibiotics. The results highlight the importance of considering both the bacterial strain and the specific antibiotic class for effective treatment strategies. Multiple antibiotic resistance halophilic were also reported earlier, suggesting that drug resistance was no more restricted to pathogenic/hospital setting bacteria, but it has wide spread in marine natural environment (Shinde et al., 2016). Interestingly, the strains that exhibited higher halotolerance, such as S. cohnii strain F and J. huakuii strain 3.7, also displayed unique protein expression patterns and varied antibiotic susceptibility profiles. The adaptive strategies observed in growth behavior and protein expression likely contribute to the strains’ ability to thrive in specific saline environments and influence their response to antibiotics. Understanding these interconnected responses is crucial for a holistic perspective on bacterial adaptation. The findings not only contribute to basic microbial physiology knowledge but also have implications for biotechnological applications and clinical considerations, especially in the context of antibiotic treatment in saline environments. ConclusionIn conclusion, our multifaceted exploration of the four-bacterial strains offers a basic understanding of their adaptive responses to hypersaline conditions. The intricate interplay between growth behavior, protein expression, and antibiotic susceptibility provides valuable insights into the unique strategies employed by these strains for survival and proliferation. The total proteins unveiled a general trend of decreasing protein content with increasing salt concentrations, with notable exceptions, such as Staphylococcus cohnii strain F, which exhibited distinct protein expression associated with ectoine biosynthesis. The expression of ectoine biosynthesis proteins highlights a potential mechanism for coping with high salinity. The antibiotic susceptibility testing results further showed the diversity in resistance profiles among the strains, emphasizing the strain-specific nature of responses to different antibiotic classes. These findings contribute not only to our fundamental understanding of microbial physiology but also hold industrial implications for ectoine production. AcknowledgmentThe authors are thankful to the Department of Microbiology, Assam Don Bosco University for providing the laboratory facility. FundingThe author declare that no funding was received for the current research. Conflict of interestThe authors declare that they have no conflict of interest. ReferencesAlam, M.M., Ahmed, R., Amin, A.M. and Hossain, N.M. 2021. Development and validation of a novel customized medium for the marine microbial culture. Biores. Commun. 7(2), 999–1003. Boltyanskaya, Yu.V., Detkova, E.N., Shumskii, A.N., Dulov, L.E. and Pusheva, M.A. 2005. Osmoadaptation in representatives of Haloalkaliphilic bacteria from Soda Lakes. Microbiol. 74(6), 640–645. https://doi.org/10.1007/s11021-005-0117-5 Choudhury, M.G. and Saha, N. 2012. Expression of inducible nitric oxide synthase and nitric oxide production in the Mud-Dwelled air-breathing Singhi Catfish, Heteropneustes Fossilis under Condition of Water Shortage. Nitric Oxide. 27(4), 219–227. https://doi.org/10.1016/j.niox.2012.07.006 Dutta, B. and Bandopadhyay, R. 2022. Biotechnological potentials of halophilic microorganisms and their impact on mankind. BJBAS. 11(1):75. https://doi.org/10.1186/s43088-022-00252-w Da Costa, P., Loureiro, L. and Matos, A. 2013. Transfer of multidrug-resistant bacteria between intermingled ecological Niches: the interface between humans, animals and the environment. Int. J. Environ. Res. Public Health. 10(1), 278–294. https://doi.org/10.3390/ijerph10010278 Das, A., Paul, S. and Choudhury, M. 2019. Isolation and molecular characterization of halophilic bacteria from Taramumbari, Coastal Region of Maharashtra, India. IJRAR. 6(2) https://ijrar.org/viewfull.php?&p_id=IJRAR19L1220 Diba, H., Cohan, R.A., Salimian, M., Mirjani, R., Soleimani, M. and Khodabakhsh, F. 2021. Isolation and characterization of halophilic bacteria with the ability of heavy metal bioremediation and nanoparticle synthesis from Khara Salt Lake in Iran. Arch. Microbiol. 203(7), 3893–3903. https://doi.org/10.1007/s00203-021-02380-w Fatollahi, P., Ghasemi, M., Yazdian, F. and Sadeghi, A. 2020. Ectoine production in bioreactor by Halomonas Elongata DSM2581: using MWCNT and Fe-Nanoparticle. Biotechnol. Prog. 37(1). https://doi.org/10.1002/btpr.3073 Fouz, N., Pangesti, K.N.A., Yasir, M., Al-Malki, A.L., Azhar, E.I., Hill-Cawthorne, G.A. and Abd El Ghany, M. 2020. The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings. IDTM. 5(1), 33. https://doi.org/10.3390/tropicalmed5010033 Galinski, E.A. 1993. Compatible solutes of Halophilic Eubacteria: molecular principles, water-solute interaction, stress protection. Experientia. 49(6-7), 487–496. https://doi.org/10.1007/bf01955150 Kang, J.Y., Lee, B., Kim, J.A., Kim, M.S. and Kim, C.H. 2022. Identification and characterization of an Ectoine Biosynthesis gene cluster from Aestuariispira Ectoiniformans Sp. Nov., isolated from seawater. Microbiol. Res. 254, 126898. https://doi.org/10.1016/j.micres.2021.126898 Mehanni, M.M., Gadow, S.I., Alshammari, F.A., Modafer, Y., Ghanem, K.Z., Noha Fathy El-Tahtawi, El-Homosy, R.F. and Hesham, A.E.L. 2023. Antibiotic-resistant bacteria in Hospital wastewater treatment plant effluent and the possible consequences of its reuse in agricultural irrigation. Front. microbial. 14, 1141383. https://doi.org/10.3389/fmicb.2023.1141383 Nakayama, H., Yoshida, K., Ono, H., Murooka, Y. and Shinmyo, A. 2000. Ectoine, the compatible solute of Halomonas Elongata, Confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiol. 122(4), 1239–1248. https://doi.org/10.1104/pp.122.4.1239 Oren, A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. JIMB. 28(1), 56–63. https://doi.org/10.1038/sj/jim/7000176 Rathakrishnan, D. and Gopalan, A.K. 2022. Isolation and characterization of halophilic isolates from Indian Salterns and their screening for production of hydrolytic enzymes. Environ. Chall. 6, 100426. https://doi.org/10.1016/j.envc.2021.100426 Rohim, R.A.A., Ahmad, W.M.A.W., Ismail, N.H., Ghazali, F.M.M., Alam, M.K. 2020. Modeling the growth of bacteria streptococcus sobrinus using exponential regression. Pesqui. Bras. Odontopediatria. Clín. Integr. 20, e5380. Shinde, V. and Thombre, R. 2016. Antibiotic resistance profiling of marine halophilic bacteria and haloarchaea. J. Appl. Pharm. Sci. 6(10), 132–137. https://doi.org/10.7324/japs.2016.601018 Thombre, R.S., Shinde, V., Thaiparambil, E., Zende, S. and Mehta, S. 2016. Antimicrobial activity and mechanism of inhibition of silver nanoparticles against extreme halophilic Archaea. Front. Microbial. 7, 1424. https://doi.org/10.3389/fmicb.2016.01424 Vaidya, V. and Manzoor, M. 2009. Horizontal transfer of antibiotic resistance in the marine environment. Nat. Environ. Pollut. Technol. 8(3), 451–458. https://neptjournal.com/upload-images/NL-5-8-(8)B-1344.pdf Vaidya, V.K. 2011. Horizontal transfer of antimicrobial resistance by extended-spectrum β Lactamase-producing Enterobacteriaceae. J. Lab. Physicians. (1), 37–42. https://doi.org/10.4103/0974-2727.78563 Wei, Y.H., Yuan, F.W., Chen, W.C. and Chen, S.Y. 2011. Production and characterization of Ectoine by Marinococcus Sp. ECT1 isolated from a high-salinity environment. JBB. 111(3), 336–342. https://doi.org/10.1016/j.jbiosc.2010.11.009 | ||
| How to Cite this Article |
| Pubmed Style Das A, Choudhury MG. Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. J Microbiol Infect Dis. 2024; 14(2): 46-54. doi:10.5455/JMID.2024.v14.i2.2 Web Style Das A, Choudhury MG. Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. https://www.jmidonline.org/?mno=192241 [Access: July 02, 2025]. doi:10.5455/JMID.2024.v14.i2.2 AMA (American Medical Association) Style Das A, Choudhury MG. Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. J Microbiol Infect Dis. 2024; 14(2): 46-54. doi:10.5455/JMID.2024.v14.i2.2 Vancouver/ICMJE Style Das A, Choudhury MG. Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. J Microbiol Infect Dis. (2024), [cited July 02, 2025]; 14(2): 46-54. doi:10.5455/JMID.2024.v14.i2.2 Harvard Style Das, A. & Choudhury, . M. G. (2024) Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. J Microbiol Infect Dis, 14 (2), 46-54. doi:10.5455/JMID.2024.v14.i2.2 Turabian Style Das, Ankur, and Mahua Gupta Choudhury. 2024. Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. Journal of Microbiology and Infectious Diseases, 14 (2), 46-54. doi:10.5455/JMID.2024.v14.i2.2 Chicago Style Das, Ankur, and Mahua Gupta Choudhury. "Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration." Journal of Microbiology and Infectious Diseases 14 (2024), 46-54. doi:10.5455/JMID.2024.v14.i2.2 MLA (The Modern Language Association) Style Das, Ankur, and Mahua Gupta Choudhury. "Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration." Journal of Microbiology and Infectious Diseases 14.2 (2024), 46-54. Print. doi:10.5455/JMID.2024.v14.i2.2 APA (American Psychological Association) Style Das, A. & Choudhury, . M. G. (2024) Growth pattern, Protein quantification, and Antibiotic responses of four Halophilic Bacterial Strains in different salt concentration. Journal of Microbiology and Infectious Diseases, 14 (2), 46-54. doi:10.5455/JMID.2024.v14.i2.2 |